 |
IMP Tutorial
for IMP version 2.4.0
|
 |
IMP Tutorial
for IMP version 2.4.0
|
In this stage, we find all available experimental data that we wish to utilize in structural modeling. In theory, any method that provides information about absolute or relative structural information can be used. A non-exhaustive list of data types currently supported in IMP is:
The rnapolii/data folder in the tutorial input files contains the data included in this example:
.mrc, .txt files)FASTA File Each residue included in modeling must be explicitly defined in the FASTA text file. Each individual component (i.e., a protein chain) is identified by a string in the FASTA header line. From 1WCM.fasta.txt:
>1WCM:A MVGQQYSSAPLRTVKEVQFGLFSPEEVRAISVAKIRFPETMDETQTRAKIGGLNDPRLGSIDRNLKCQTCQEGMNECPGH FGHIDLAKPVFHVGFIAKIKKVCECVCMHCGKLLLDEHNELMRQALAIKDSKKRFAAIWTLCKTKMVCETDVPSEDDPTQ ... >1WCM:B MSDLANSEKYYDEDPYGFEDESAPITAEDSWAVISAFFREKGLVSQQLDSFNQFVDYTLQDIICEDSTLILEQLAQHTTE SDNISRKYEISFGKIYVTKPMVNESDGVTHALYPQEARLRNLTYSSGLFVDVKKRTYEAIDVPGRELKYELIAEESEDDS ...
defines two chains with unique IDs of 1WCM:A and 1WCM:B respectively. The entire complex is 12 chains and 4582 residues.
Electron Density Map The electron density map of the entire RNA Poly II complex is at 20.9 Angstrom resolution. The raw data file for this is stored in emd_1883.map.mrc.

Electron microscopy density map for yeast RNA Polymerase II
Electron Density as Gaussian Mixture Models Gaussian mixture models (GMMs) are used to greatly speed up scoring by approximating the electron density of individual subunits and experimental EM maps. A GMM has been created for the experimental density map, and is stored in emd_1883.map.mrc.gmm.50.mrc. The weight, center, and covariance matrix of each Gaussian used to approximate the original EM density can be seen in the corresponding .txt file.
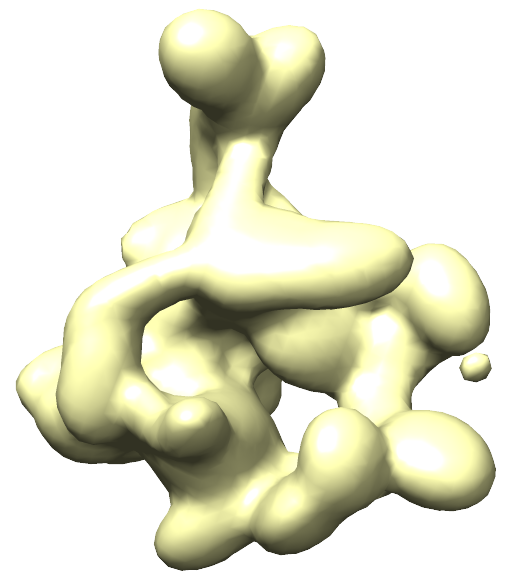
The EM data represented as a 50 Gaussian mixture model
PDB File High resolution coordinates for all 12 chains of RNA Pol II are found in 1WCM.pdb.
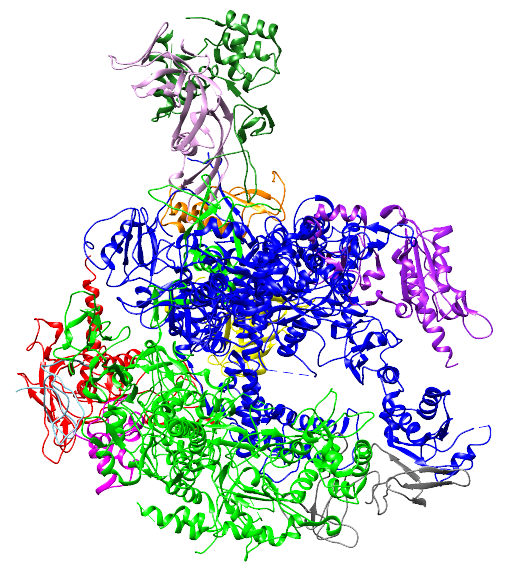
Coordinates from PDBID 1WCM
Chemical Cross-Links All chemical cross-linking data is located in polii_xlinks.csv and polii_juri.csv. These files contain multiple comma-separated columns; four of these specify the protein and residue number for each of the two linker residues.
prot1,res1,prot2,res2 Rpb1,34,Rpb1,49 Rpb1,101,Rpb1,143 Rpb1,101,Rpb1,176
The length of the DSS/BS3 cross-linker reagent, 21 angstroms, will be specified later in the modeling script.
With data gathered, we can now proceed to Stage 2 - Representation of subunits and translation of the data into spatial restraints.