|
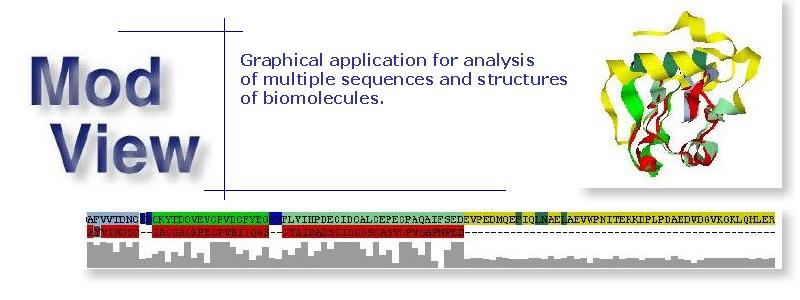
ModView is a program to visualize and analyze multiple biomolecule structures and/or sequence
alignments. As a Netscape plugin,like
Chime
, it can be embed into Web pages and controlled by
Javascript objects on the page.
It has wide range of tools to manipulate and analyze sequences and structures by interactive control
... ModView as Front-end interface to biodatabases
Modview can easily interface with
local or remote bioresources to provide GUI and analytical tools. Currently
ModView is used as a database interface in several structure-sequence protein
resources.
ModBase,
a Database of Comparative Protein Structure Models, (Pieper et. al, Nuc.Acid.Res.)
LigBase,
a Structural Database of Aligned Ligand Binding Protein Sequences, (Stuart et al., Bioinformatics).
DBAli,
a Database of Sequence-Structure alignments, (Marti-Renom et. al.2001,Bioinformatics).
ModView can be also used to view structures and sequences
in
Protein Data Bank ,
choose 'Rasmol' display option on View Structure window.
... ModView provides Protein Family analysis.
Two families of DNA binding proteins have been analysed by ModView.
The scalable multiple alignment with combination of
visualization of structural features of molecules cleary shows invariant residues
involved in DNA binding in
Zinc finger protein superfamily (ZFP) and superfamily of the DNA-Binding Domain
Of The Replication Initiation Protein E1 From Papillomavirus (RIP). These two families
also shows different pictures of homology tree.
ZFP family has ~50% identity among all the members,
while RIP family can be subdivided in several closely related subfamilies.
(Ilyin and Sali, 2001, CSHL).
Recently, ModView facilitates the target selection process for
New York Structural Genomics Research
Consortium, by providing integrative visual analyses of the comparative
protein models, PSI-BLAST profile and phylogenetic data of protein targets
selected for novel structures determination (Bonanno et. al. 2001, PNAS).
... ModView References.
* V.A.Ilyin, U. Pieper, A. C. Stuart, M. A. Mart-Renom and A. Sali.
Analysis and Visualization of Multiple Biomolecule Structures and
Sequences by ModView. (submitted)
* Ursula Pieper, Narayanan Eswar, Ashley C. Stuart, Valentin A. Ilyin and
Andrej Sali, MODBASE, a database of annotated comparative protein
structure models. Nucleic Acids Research. 2002. Jan 1: 30(1):255-9.
PubMed
* Stuart A.C., Ilyin V.A., and Sali A., (2001) LigBase: A Database of
Families of Aligned Ligand Binding Sites in Known Protein Sequences and
Structures, Bioinformatics 2002 Jan;18(1):200-1 PubMed
* Jeffrey B. Bonanno, Carme Edo, Narayanan Eswar, Urusla Pieper, Valentin
Ilyin, Michael J. Romanowski, Sue Ellen Gerhaman, Helen Kycia, F. William
Sudier, Andrej Sali, Stephen Burley. Structural genomics of enzymes
involved in sterol/isoprenoid biosynthesis. Proc Natl Acad Sci U S A 2001 Nov 6;98(23):12896-901.
PubMed
*
Valentin Ilyin and Andrej Sali. Protein superfamily analysis by ModView.
CSHL meeting. Aug.8, 2001.
* Marti-Renom, M.A., Ilyin V.A., Sali, A. DBAli: A database of protein
structure alignments. Bioinformatics. p.746. Vol. 17 no 8, 2001.
PubMed
Modview uses Roger Sayle's
Rasmol rendering system and
modified alignment editor panel from Michele Clamp's
Jalview.
|