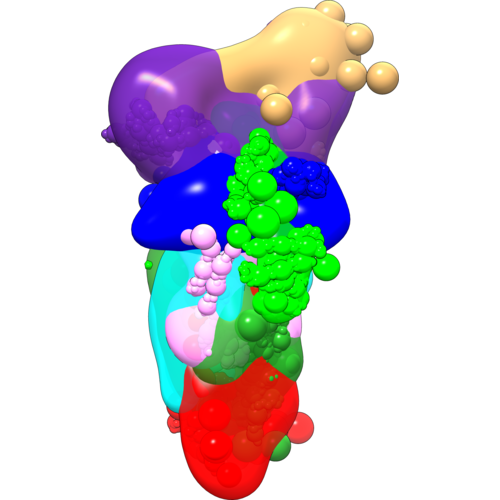
These scripts demonstrate the use of IMP in the modeling of the yeast exocyst complex using DSS and EDC datasets, and a negative-stain EM density map. The scripts work with the IMP (Release 2.8.0).
datacontains all relevant dataproduction_scriptcontains all the modeling scripts version for the different setups.analysis_scriptscontains all the analysis scripts.archivingcontains the cif file and script used to generate itresultscontains the models, centroid structure and localization densities
Author(s): Sai Ganesan
License: CC BY-SA 4.0 This work is licensed under the Creative Commons Attribution-ShareAlike 4.0 International License.
Publications: Ganesan SJ, Feyder MJ, Chemmama IE, et al. Integrative structure and function of the yeast exocyst complex. Protein Sci. 2020;29(6):1486-1501. doi:10.1002/pro.3863



 Download files
Download files Verified to work with the
Verified to work with the  To install the software needed to reproduce this system with the
To install the software needed to reproduce this system with the
 To set up the environment on the UCSF Wynton cluster to run
this system, run:
To set up the environment on the UCSF Wynton cluster to run
this system, run: