FILE =
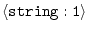 |
'default' |
name of the coordinates' file |
ATOM_FILES_DIRECTORY =
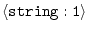 |
'./' |
input atom files directory list (e.g., 'dir1:dir2:dir3:./:/') |
MODEL2_SEGMENT =
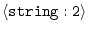 |
'FIRST:@' 'LAST:' |
segment to be read in |
MODEL_FORMAT =
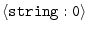 |
'PDB' |
selects input atom file format: 'PDB' | 'CHARMM' | 'UHBD' |
WATER_IO =
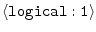 |
off |
whether to read water coordinates |
HETATM_IO =
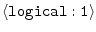 |
off |
whether to read HETATM coordinates |
HYDROGEN_IO =
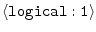 |
off |
whether to read hydrogen coordinates |