| io = <io_data> |
|
Options for reading atom files |
| rr_file = <str:1> |
'$(LIB)/as1.sim.mat' |
input residue-residue scoring file |
| gap_penalties_1d = <float:2> |
900 50 |
gap creation and extension penalties for sequence/sequence alignment |
| gap_penalties_2d = <float:9> |
0.35 1.2 0.9 1.2 0.6 8.6 1.2 0 0 |
gap penalties for sequence/structure alignment: helix, beta, accessibility, straightness, and CA-CA distance factor, dst min, dst power, t, structure_profile ; best U,V=-450,0 |
| align_block = <int:1> |
0 |
the last sequence in the first block of sequences |
| max_gap_length = <int:1> |
999999 |
maximal length of gap in protein comparisons |
| off_diagonal = <int:1> |
100 |
to speed up the alignment |
| matrix_offset = <float:1> |
0.00 |
substitution matrix offset for local alignment |
| overhang = <int:1> |
0 |
un-penalized overhangs in protein comparisons |
| local_alignment = <bool:1> |
False |
whether to do local as opposed to global alignment |
| align_what = <str:1> |
'BLOCK' |
what to align in ALIGN; 'BLOCK' | 'ALIGNMENT' | 'LAST' | 'PROFILE' |
| subopt_offset = <float:1> |
0.0 |
offset for residue-residue score in getting suboptimals in ALIGN/ALIGN2D |
| fit = <bool:1> |
True |
whether to align |
| read_weights = <bool:1> |
False |
whether to read the whole NxM weight matrix for ALIGN* |
| write_weights = <bool:1> |
False |
whether to write the whole NxM weight matrix for ALIGN* |
| input_weights_file = <str:1> |
'' |
Exteral weight matrix input to MODELLER (SALIGN/ALIGN) |
| output_weights_file = <str:1> |
'' |
File into which the weight file is wriiten (iff WRITE_WEIGHTS = 'on') |
| weigh_sequences = <bool:1> |
False |
whether or not to weigh sequences in a profile |
| smooth_prof_weight = <float:1> |
10 |
for smoothing the profile aa frequency with a prior |
| read_profile = <bool:1> |
False |
whether to read str profile for ALIGN2D |
| input_profile_file = <str:1> |
'' |
multiple sequece alignment read into MODELLER for profile-profile alignments |
| write_profile = <bool:1> |
False |
whether to write str profile for ALIGN2D |
| output_profile_file = <str:1> |
'' |
|
- Description:
- This command aligns a block of sequences (second block)
with a block of structures (first block). It is the same as the
alignment.align() command except that a variable gap opening penalty is
used. This gap penalty depends on the 3D structure of all sequences in
block 1. The variable gap penalty can favor gaps in exposed regions,
avoid gaps within secondary structure elements, favor gaps in
curved parts of the mainchain, and minimize the distance between the two
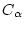 positions spanning a gap. The alignment.align2d() command is
preferred for aligning a sequence with structure(s) in comparative
modeling because it tends to place gaps in a better structural
context. See Section 5.1.2 for the dynamic
programming algorithm that implements the variable gap penalty.
gap_penalties_2d specifies parameters
positions spanning a gap. The alignment.align2d() command is
preferred for aligning a sequence with structure(s) in comparative
modeling because it tends to place gaps in a better structural
context. See Section 5.1.2 for the dynamic
programming algorithm that implements the variable gap penalty.
gap_penalties_2d specifies parameters
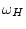 ,
, 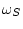 ,
, 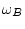 ,
, 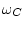 ,
, 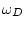 ,
,
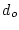 ,
, 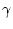 ,
, 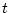 and
and 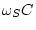 . (Section 5.1.2).
The default gap penalties gap_penalties_1d (
. (Section 5.1.2).
The default gap penalties gap_penalties_1d (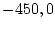 )
and gap_penalties_2d (0.35, 1.2, 0.9, 1.2, 0.6, 8.6, 1.2, 0.0, 0.0)
as well as the rr_file substitution
matrix ('as1.sim.mat') were found to be optimal in pairwise alignments of
structures and sequences sharing from 30% to 45% sequence identity
(MSM, MAM-R, RS and AŠ, in preparation).
)
and gap_penalties_2d (0.35, 1.2, 0.9, 1.2, 0.6, 8.6, 1.2, 0.0, 0.0)
as well as the rr_file substitution
matrix ('as1.sim.mat') were found to be optimal in pairwise alignments of
structures and sequences sharing from 30% to 45% sequence identity
(MSM, MAM-R, RS and AŠ, in preparation).
-- move to back
The linear gap penalty function for inserting a gap in block 1 of
structures is:
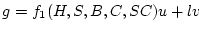 where
where 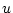 and
and 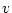 are the
usual gap opening and extension penalties,
are the
usual gap opening and extension penalties, 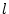 is gap length, and
is gap length, and
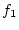 is a function that is at least 1, but can be larger to make gap
opening more difficult in the following circumstances: between two consecutive
(i.e.,
is a function that is at least 1, but can be larger to make gap
opening more difficult in the following circumstances: between two consecutive
(i.e., 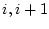 ) helical positions, two consecutive
) helical positions, two consecutive 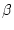 -strand positions, two
consecutive buried positions, or two consecutive positions where
the mainchain is locally straight. This function is
-strand positions, two
consecutive buried positions, or two consecutive positions where
the mainchain is locally straight. This function is
![$f_1 = 1 + [\omega_H
H_i H_{i+1} + \omega_S S_i S_{i+1} + \omega_B B_i B_{i+1} +
\omega_C C_i C_{i+1} + \omega_SC SC_i SC_{i+1}]$](img143.png) ,
, 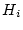 is the fraction of
helical residues at position
is the fraction of
helical residues at position 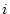 in block 1,
in block 1, 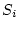 is the fraction of
is the fraction of
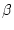 -strand residues at position
-strand residues at position 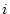 in block 1,
in block 1, 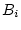 is the average relative
sidechain buriedness of residues at position
is the average relative
sidechain buriedness of residues at position 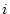 in block 1,
in block 1, 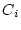 is the
average straightness of residues at position
is the
average straightness of residues at position 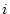 in block 1, and
in block 1, and 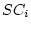 is the
strucutural conserveredness at position
is the
strucutural conserveredness at position 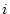 in block 1. See
Section 3.6.24 for the definition of these features.
The original straightness is modified here by assigning maximal straightness
of 1 to all residues in a helix or a
in block 1. See
Section 3.6.24 for the definition of these features.
The original straightness is modified here by assigning maximal straightness
of 1 to all residues in a helix or a 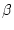 -strand. The structural conservedness
of the residues in block 1 are imported from an external source
"input_profile_file". The structural conservedness at a particular position
gives the liklehood of the occurance of a gap when structurally similar
regions from all know protein structures are aligned structurally.
-strand. The structural conservedness
of the residues in block 1 are imported from an external source
"input_profile_file". The structural conservedness at a particular position
gives the liklehood of the occurance of a gap when structurally similar
regions from all know protein structures are aligned structurally.
The linear gap penalty function for opening a gap in block 2 of sequences
is:
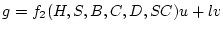 where
where 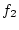 is a
function that is at least 1, but can be larger to make the gap
opening in block 2 more difficult in the following circumstances:
when the first gap position is aligned with a helical residue, a
is a
function that is at least 1, but can be larger to make the gap
opening in block 2 more difficult in the following circumstances:
when the first gap position is aligned with a helical residue, a 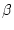 -strand
residue, a buried residue, extended mainchain, or when the whole gap in
block 2 is spanned by two residues in block 1 that are far apart in space.
This function is
-strand
residue, a buried residue, extended mainchain, or when the whole gap in
block 2 is spanned by two residues in block 1 that are far apart in space.
This function is
![$f_2 = 1 + [\omega_H H_i + \omega_S S_i + \omega_B B_i +
\omega_C C_i + \omega_D \sqrt{d-d_o} + \omega_SC SC_i]$](img150.png) .
. 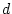 is the distance
between the two
is the distance
between the two 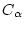 atoms spanning the gap, averaged over all structures
in block 1 and
atoms spanning the gap, averaged over all structures
in block 1 and 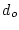 is the distance that is small enough to correspond to no
increase in the opening gap penalty (e.g., 8.6).
is the distance that is small enough to correspond to no
increase in the opening gap penalty (e.g., 8.6).
When fit is False, no alignment is done and the routine returns
only the average structural information, which can be written out by the
alignment.write() command.
- Example: examples/commands/align2d.py
-
# Demonstrating ALIGN2D, aligning with variable gap penalty
log.verbose()
env = environ()
env.libs.topology.read(file='$(LIB)/top_heav.lib')
# Read aligned structure(s):
aln = alignment(env)
aln.append(file='toxin.ali', align_codes='2ctx')
aln_block = len(aln)
# Read aligned sequence(s):
aln.append(file='toxin.ali', align_codes='1nbt')
# Structure sensitive variable gap penalty sequence-sequence alignment:
aln.align2d(overhang=0, gap_penalties_1d=(-450, 0),
gap_penalties_2d=(0.35, 1.2, 0.9, 1.2, 0.6, 8.6, 1.2, 0., 0.),
align_block=aln_block)
aln.write(file='align2d.ali', alignment_format='PIR')
aln.write(file='align2d.pap', alignment_format='PAP',
alignment_features='INDICES HELIX BETA STRAIGHTNESS ' + \
'ACCESSIBILITY CONSERVATION')
aln.check()
# Color the first template structure according to gaps in alignment:
aln = alignment(env)
aln.append(file='align2d.ali', align_codes=('2ctx', '1nbt'),
alignment_format='PIR', remove_gaps=True)
mdl = model(env)
mdl.read(aln=aln, model_segment=('2ctx', '2ctx'))
mdl.color(aln=aln)
mdl.write(file='2ctx.aln.pdb')
# Color the first template structure according to secondary structure:
mdl.write_data(file='2ctx', output='SSM')
mdl.write(file='2ctx.ssm.pdb')
# Superpose the target structure onto the first template:
mdl2 = model(env)
mdl2.read(aln=aln, model_segment=('1nbt', '1nbt'))
mdl.pick_atoms(aln=aln, atom_types='CA')
mdl.superpose(mdl2, aln)
mdl2.write(file='1nbt.fit.pdb')
![]() where
where ![]() and
and ![]() are the
usual gap opening and extension penalties,
are the
usual gap opening and extension penalties, ![]() is gap length, and
is gap length, and
![]() is a function that is at least 1, but can be larger to make gap
opening more difficult in the following circumstances: between two consecutive
(i.e.,
is a function that is at least 1, but can be larger to make gap
opening more difficult in the following circumstances: between two consecutive
(i.e., ![]() ) helical positions, two consecutive
) helical positions, two consecutive ![]() -strand positions, two
consecutive buried positions, or two consecutive positions where
the mainchain is locally straight. This function is
-strand positions, two
consecutive buried positions, or two consecutive positions where
the mainchain is locally straight. This function is
![]() ,
, ![]() is the fraction of
helical residues at position
is the fraction of
helical residues at position ![]() in block 1,
in block 1, ![]() is the fraction of
is the fraction of
![]() -strand residues at position
-strand residues at position ![]() in block 1,
in block 1, ![]() is the average relative
sidechain buriedness of residues at position
is the average relative
sidechain buriedness of residues at position ![]() in block 1,
in block 1, ![]() is the
average straightness of residues at position
is the
average straightness of residues at position ![]() in block 1, and
in block 1, and ![]() is the
strucutural conserveredness at position
is the
strucutural conserveredness at position ![]() in block 1. See
Section 3.6.24 for the definition of these features.
The original straightness is modified here by assigning maximal straightness
of 1 to all residues in a helix or a
in block 1. See
Section 3.6.24 for the definition of these features.
The original straightness is modified here by assigning maximal straightness
of 1 to all residues in a helix or a ![]() -strand. The structural conservedness
of the residues in block 1 are imported from an external source
"input_profile_file". The structural conservedness at a particular position
gives the liklehood of the occurance of a gap when structurally similar
regions from all know protein structures are aligned structurally.
-strand. The structural conservedness
of the residues in block 1 are imported from an external source
"input_profile_file". The structural conservedness at a particular position
gives the liklehood of the occurance of a gap when structurally similar
regions from all know protein structures are aligned structurally.
![]() where
where ![]() is a
function that is at least 1, but can be larger to make the gap
opening in block 2 more difficult in the following circumstances:
when the first gap position is aligned with a helical residue, a
is a
function that is at least 1, but can be larger to make the gap
opening in block 2 more difficult in the following circumstances:
when the first gap position is aligned with a helical residue, a ![]() -strand
residue, a buried residue, extended mainchain, or when the whole gap in
block 2 is spanned by two residues in block 1 that are far apart in space.
This function is
-strand
residue, a buried residue, extended mainchain, or when the whole gap in
block 2 is spanned by two residues in block 1 that are far apart in space.
This function is
![]() .
. ![]() is the distance
between the two
is the distance
between the two ![]() atoms spanning the gap, averaged over all structures
in block 1 and
atoms spanning the gap, averaged over all structures
in block 1 and ![]() is the distance that is small enough to correspond to no
increase in the opening gap penalty (e.g., 8.6).
is the distance that is small enough to correspond to no
increase in the opening gap penalty (e.g., 8.6).