- ... alignment.2.1
- If the residue type is defined in
'modlib/restyp.lib' you can use the 1-letter code that is specified there,
but if in doubt use '.', since that matches everything.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... assigned2.2
- After
uppercase letters A-Z are used, chain IDs 0 through 9 are assigned, then
lowercase letters a-z. If your protein contains more than 62 chains, the
remaining chains are given no IDs.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... restraints2.3
- The
restraints include bond, angle and dihedral angle restraints.
The SG — SG atom pair also becomes an excluded atom pair that is not checked
for an atom-atom overlap. The
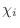 dihedral angle restraints will depend
on the conformation of the equivalent disulfides in the template structure,
as described in [Šali & Overington, 1994].
dihedral angle restraints will depend
on the conformation of the equivalent disulfides in the template structure,
as described in [Šali & Overington, 1994].
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ...
exception5.1
- MODELLER can raise the following Python exceptions:
ZeroDivisionError, IOError, MemoryError, EOFError,
TypeError, NotImplementedError, UnicodeError, IndexError, and ValueError.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... special.5.2
- On Unix/Linux systems, MODELLER assumes that your filesystem
also stores filenames in UTF-8. This is usually the case on modern systems;
however, you can change the encoding by setting the G_FILENAME_ENCODING
environment variable.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... numbers6.1
- for mmCIF format,
author-provided residue numbers (_atom_site.auth_seq_id) are used if
available rather than the canonical sequence id
(_atom_site.label_seq_id)
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... library6.2
- If no atoms in the first
chain have coordinates, internal coordinate generation is seeded by placing the
first atom at the origin, the second along the x axis, and the third in the xy
plane. If no atoms in subsequent chains have coordinates, the first atom is
placed 2 angstroms along each of the x, y and z axes from the last atom in the
previous chain, and the second and third atoms placed in the same way as for
the first chain.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... communication)6.3
-
Fractional surface area of a residue X is given as the calculated surface area
divided by that of the residue in an extended tripeptide Ala-X-Ala
conformation.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ...
scores6.4
- The mean and standard deviation of the DOPE score of a given
protein is estimated from its sequence. The mean score of a random protein
conformation is estimated by a weighted sum of protein composition over the
20 standard amino acid residue types, where each weight corresponds to the
expected change in the score by inserting a specific type of amino acid
residue. The weights are estimated from a separate training set of
1,686,320 models generated by MODPIPE.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ...
6.5
- The target distances were all obtained from a regular α-helix in
one of the high-resolution myoglobin structures.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ...http://www.ks.uiuc.edu/Research/vmd/.
6.6
- Note that binary trajectory files are machine dependent; it is up
to the visualization software to do any necessary byte-swapping.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... files6.7
- This won't work in combination
with write_whole_pdb = False for structures
that were added to the alignment with alignment.append_model(), since
such inputs may not have an atom file to extract the directory from. In this
case the outputs will end up in the current directory.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ...
out6.8
- Any structures that were added with alignment.append_model()
will need to have their corresponding atom files available, so that the
originals can be reread at this point.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... files6.9
- This won't work in combination
with write_whole_pdb = False for structures
that were added to the alignment with alignment.append_model(), since
such inputs may not have an atom file to extract the directory from. In this
case the outputs will end up in the current directory.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ...
out6.10
- Any structures that were added with alignment.append_model()
will need to have their corresponding atom files available, so that the
originals can be reread at this point.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ...'m'6.11
- Note that 'm.chains['A'].residues['10']' in most cases is
the same as 'm.residues['10:A']'. However, if there are multiple chains in
the model called 'A' the first syntax will return residue 10 in the first chain
called A (and so will fail if that chain does not contain this residue) whereas
the second will look for the first residue numbered 10 in any of the A chains.
(Generally speaking, you should avoid having duplicate chain IDs or residue
numbers!)
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... codeB.1
-
for PDB format, this is a 1-letter chain ID; for mmCIF format, this is the
author-provided chain/asym ID (_atom_site.auth_asym_id) if available,
otherwise the canonical asym ID (_atom_site.label_asym_id), both of
which can be multiple characters in length
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.