



Next: alignment.check_sequence_structure() check
Up: The alignment class: comparison
Previous: alignment.read_one() read
Contents
Index
check_structure_structure(eqvdst=6.0, io=None)
- Output:
- n_exceed
This command checks the alignment of the template structures
(all but the last entry in the alignment):
For each pairwise superposition of the templates,
it reports those equivalent pairs of
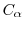 atoms that are
more than eqvdst Å away from each other. Such pairs are almost
certainly misaligned. The pairwise superpositions are done using the
atoms that are
more than eqvdst Å away from each other. Such pairs are almost
certainly misaligned. The pairwise superpositions are done using the
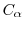 atoms and the given alignment. The number of such pairs is returned.
atoms and the given alignment. The number of such pairs is returned.
Note that the target structures are actually changed by the superpositions
carried out by this command. If you want to use these superpositions as a
crude initial model for automodel model building (rather than
setting automodel.initial_malign3d) please bear in mind that the later
templates in your alignment are always fitted on the earlier templates. Thus,
a more reliable initial model will be obtained if you list the higher coverage
templates earlier in the knowns variable in your automodel script
(e.g., always list multimeric templates before monomers).
If you want to use the original non-superposed structures, either
avoid calling this command, or delete and recreate the alignment object
afterwards to force it to reread the structure files.




Next: alignment.check_sequence_structure() check
Up: The alignment class: comparison
Previous: alignment.read_one() read
Contents
Index
Automatic builds
2008-10-07
atoms that are more than eqvdst Å away from each other. Such pairs are almost certainly misaligned. The pairwise superpositions are done using the
atoms and the given alignment. The number of such pairs is returned.