



Next: automodel.special_patches() add
Up: automodel reference
Previous: automodel.special_restraints() add
Contents
Index
nonstd_restraints(aln)
This routine adds restraints to keep residues that have no defined topology
(generally BLK residues, used to transfer ligands directly from templates,
as described in Section 2.2.1) in a reasonable
conformation. You can override this method if you need to change these
restraints.
By default, four sets of restraints are built:
- Intra-residue distances are all constrained to their template values.
This causes each residue to behave as a rigid body.
- Inter-residue distances are constrained to template values if these are
7Å or less. This has the effect of preserving multiple-HETATM
structures such as DNA chains. If the distances are 2.3Å or less they
are assumed to be bonds and so are also excluded from the nonbonded list.
- Residue-protein atom distances are constrained to template values (and
excluded from the nonbonded list) if these are 2.3Å or less. This
preserves chemical bonds between ligands and the protein.
- Residue-protein
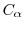 distances are constrained to template values if
these are 10Å or less and are not already bonded by the previous
restraints. This preserves the ligand position.
distances are constrained to template values if
these are 10Å or less and are not already bonded by the previous
restraints. This preserves the ligand position.




Next: automodel.special_patches() add
Up: automodel reference
Previous: automodel.special_restraints() add
Contents
Index
Automatic builds
2009-06-12