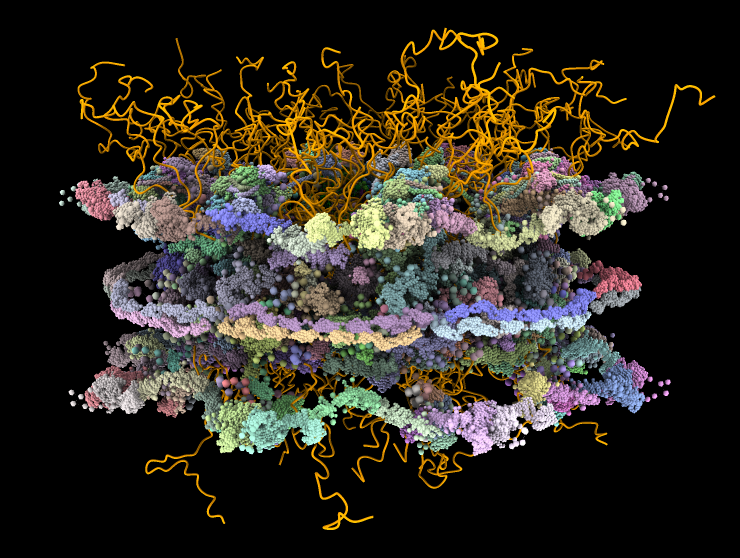
Synopsis: The files in this folder can be used to reproduce the Brownian Dynamics simulations of FG repeats using IMP as described in Kim et al., 2018
Author: Barak Raveh
E-mail: barak.raveh@gmail.com or barak@salilab.org
Date last updated: Jan 9th, 2018
Scripts/ - scripts for generating the FG repeats over an input scaffold
InputData/ - folder of input RMF file describing the scaffold
Output/ - output from running the script, as explained below
SampleOutput/ - sample of the main output from running the script
RepresentativeEnsemble/ - ensemble of output models (extracted from simulation outputs stored on Andrej's Salilab park4 file system, in folder /salilab/park4/barak/Runs/NPC_FullModel2016/FullNPC_Oct10_Cluster0model_Rg70_per_600aa_InflateObstacles_InflatedKaps, see .../Ensemble/make_ensemble.sh there). The accompanying .dcd file contains a larger number of structures from the same ensemble in CHARMM/NAMD DCD format, designed to work together with the NPC mmCIF structure deposited at PDB-Dev. It is generated using util/to_dcd.py.
Densities/ - densities of various nups and all nups, from same folder as RepresentativeEnsemble/
-
Download IMP (https://github.com/salilab/imp) - a recent nightly (or develop) build is needed
-
Build IMP according to online instructions, including the IMP.npctransport module (this requires Protobuf)
-
Create folder for output, e.g. "
Output" -
Create model of NPC from RMF file of scaffold (expected running time - a few minutes) in output folder ("
Output" in this example):$ Scripts/load_whole_new_coarse_grained_v5.py Output/config.pb InputData/47-35_1spoke.rmf3 >& Output/config.txt & -
move to
Outputfolder:$ cd Output -
Equilibrate and run for as long as desired by changing
short_init_factorandshort_sim_factorfor shorter or longer equilibration and simulation, respectively, and using a specific random seed using optional--random_seedflag; output file and movie file names could be changed as well, use--helpoption for more information (expected running time - hours to days depending on simulation time and system):$ fg_simulation --configuration config.pb --output output.pb --short_init_factor 0.25 --short_sim_factor 1.0 --conformations movie.rmf --random_seed $RANDOM >& LOG.fg_simulation & -
The output movie file
movie.rmfcan be viewed using e.g. Chimera.
Author(s): Barak Raveh
Date: March 8th, 2018
License: CC-BY-SA-4.0. This work is freely available under the terms of the Creative Commons Attribution-ShareAlike 4.0 International License.
Publications:
- Seung Joong Kim*, Javier Fernandez-Martinez*, Ilona Nudelman*, Yi Shi*, Wenzhu Zhang*, et al., Integrative structure and Functional Anatomy of a Nuclear Pore Complex, Nature 555, 475-482, 2018.



 Download files
Download files Verified to work with the
Verified to work with the  To install the software needed to reproduce this system with the
To install the software needed to reproduce this system with the
 To set up the environment on the UCSF Wynton cluster to run
this system, run:
To set up the environment on the UCSF Wynton cluster to run
this system, run: