This directory contains the input data, protocols and output model for the modeling of the PhoQ homodimer, using cysteine crosslinking and multi-state Bayesian modeling in IMP.
solution_ensemble:
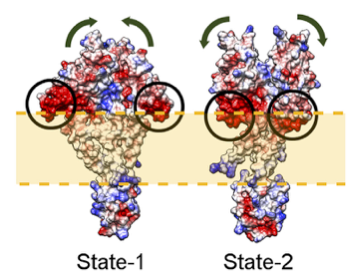
all the 2-state models within the same cluster of similar structures.
Each model is made of two structures, labeled by number 1 and 2.
localization_densities:
all the calculated localization densities, for given domains, and for the
two states.
ucsf_chimera_files:
files used to generate the figures in the paper.
modeling:
scripts for modeling the system:
2Y20_MC_1_FULL1-state modeling2Y20_MC_2_FULL2-state modeling2Y20_MC_2_FULL_90_PERCENT_DATA_12-state modeling with 90% data jackknifing2Y20_MC_2_FULL_90_PERCENT_DATA_22-state modeling with 90% data jackknifing2Y20_MC_2_FULL_95_PERCENT_DATA_12-state modeling with 95% data jackknifing2Y20_MC_2_FULL_95_PERCENT_DATA_22-state modeling with 95% data jackknifing2Y20_MC_3_FULL3-state modeling
To run a modeling script, just change into its directory and run it from the command line, e.g.
cd modeling/2Y20_MC_2_FULL./HK_model_ENSEMBLE_REM.py(on a single processor; prependmpirun -np 4or similar if you built IMP with MPI support)
Two output files will be produced: a log file containing statistics, and a trajectory of model structures in RMF format. The latter can be viewed in UCSF Chimera.
Author(s): Max Bonomi, Riccardo Pellarin, Ben Webb
Date: March 18th, 2015
License: LGPL. This library is free software; you can redistribute it and/or modify it under the terms of the GNU Lesser General Public License as published by the Free Software Foundation; either version 2 of the License, or (at your option) any later version.
Testable: Yes.
Parallelizeable: Yes
Publications:
- K. Molnar, M. Bonomi, R. Pellarin, G. Clinthorne, G. Gonzalez, S. Goldberg, M. Goulian, A. Sali, W. DeGrado. Cys-Scanning Disulfide Crosslinking and Bayesian Modeling Probe the Transmembrane Signaling Mechanism of the Histidine Kinase, PhoQ, Structure 22, 1239-1251, 2014.



 Download files
Download files Verified to work with the
Verified to work with the  To install the software needed to reproduce this system with the
To install the software needed to reproduce this system with the
 To set up the environment on the UCSF Wynton cluster to run
this system, run:
To set up the environment on the UCSF Wynton cluster to run
this system, run: