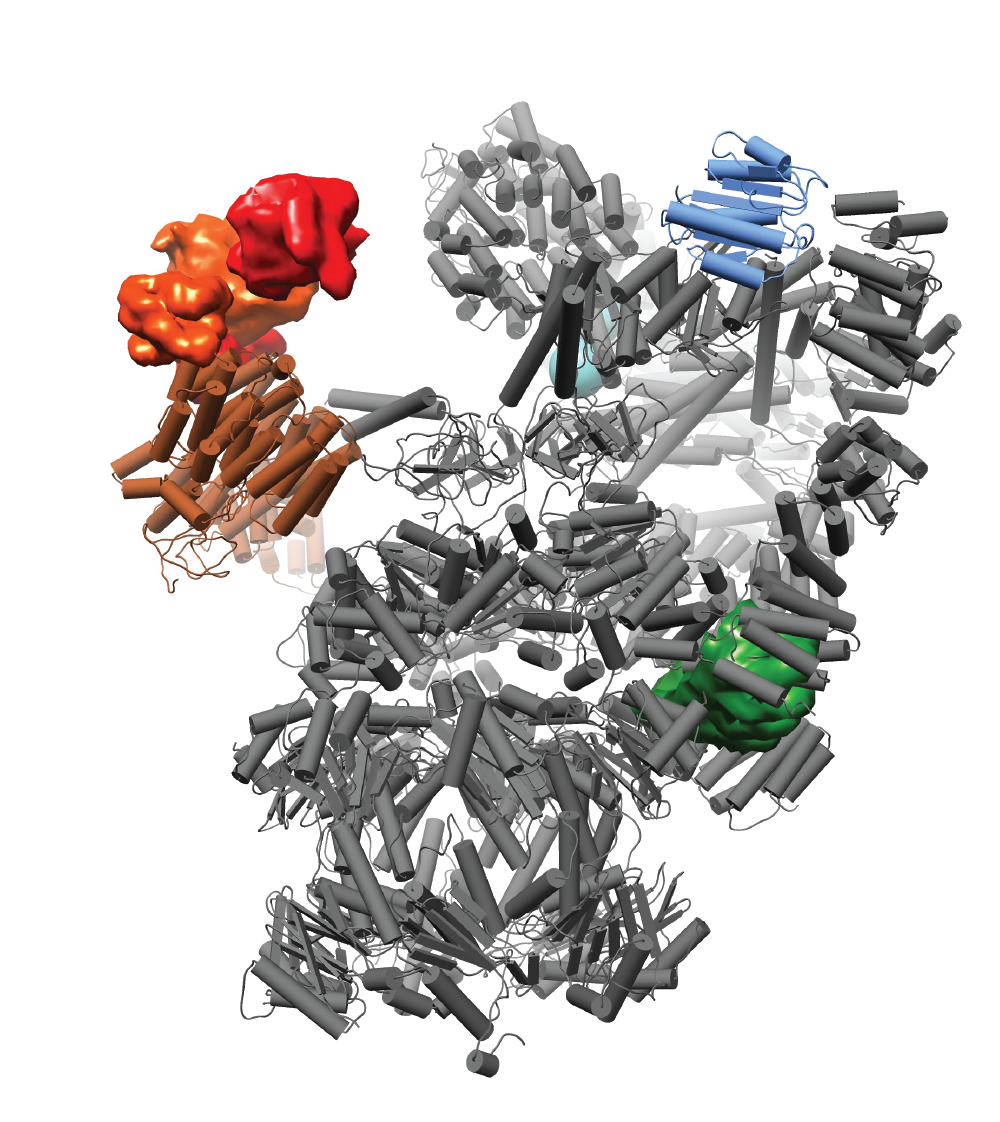
The modeling.py script demonstrates the use of IMP and PMI in the modeling of the 26S proteasome - UBLCP1 complex, using chemical crosslinking and comparative modeling.
The modeling protocol will work with a default build of IMP.
-
modeling.pythe main IMP/PMI script for modeling -
inputsh26s_flex_fix.pdbcomparative model of the 26S proteasome structureublcp1_t1_1.pdbcomparative model of the UBLCP1 structure with its Ubl domain bound to T1 siteublcp1_t1_2.pdbcomparative model of the UBLCP1 structure with its Ubl domain bound to T2 sitesequences.fastaFASTA file with all the protein sequencestopology.txtIMP/PMI topology input filexlinks.txtlist of cross-links
-
outputAn example IMP/PMI output directory obtained by running the modeling.py script
python modeling.py
Author(s): Peter Cimermancic, Charles Greenberg
Date: May 17th, 2016
License: CC-BY-SA-4.0. This work is freely available under the terms of the Creative Commons Attribution-ShareAlike 4.0 International License.
Publications:
- X. Wang, P. Cimermancic, C. Yu, E. Sakata, X. Guo, C. Greenberg, A. Schweitzer, A.S. Huszagh, Y. Yang, E.J. Novitsky, A. Leitner, P. Nanni, A. Kahraman, J. Dixon, S.D. Rychnovsky, R. Aebersold, W. Baumeister, A. Sali, L. Huang. Molecular Details Underlying Dynamic Structures and Regulation of the Human 26S Proteasome, Mol Cell Proteomics 16(5):840-854, 2017



 Download files
Download files Verified to work with the
Verified to work with the  To install the software needed to reproduce this system with the
To install the software needed to reproduce this system with the
 To set up the environment on the UCSF Wynton cluster to run
this system, run:
To set up the environment on the UCSF Wynton cluster to run
this system, run: