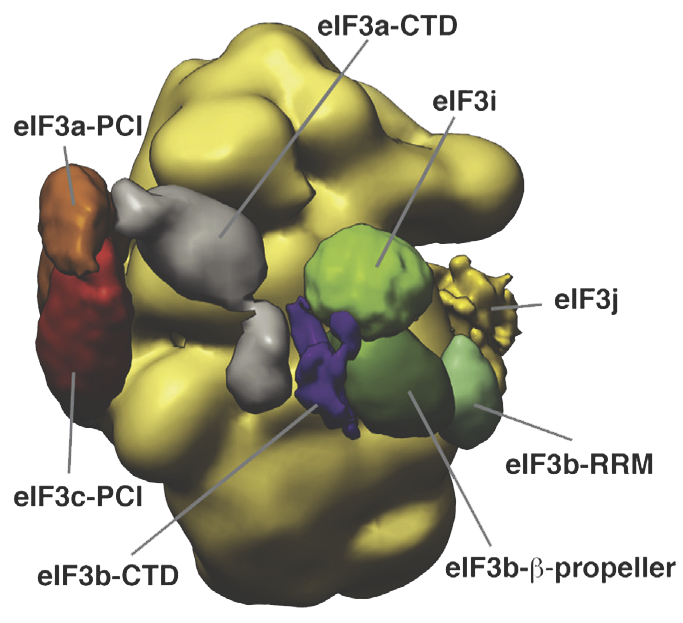
Eukaryotic translation initiation requires the recruitment of the large, multiprotein eIF3 complex to the 40S ribosomal subunit. Using X-ray structures of all major components of the minimal, six-subunit Saccharomyces cerevisiae eIF3 core, together with cross-linking coupled to mass spectrometry, we were able to use IMP to position and orient all eIF3 components on the 40S•eIF1 complex, revealing an extended, modular arrangement of eIF3 subunits.
cd modeling/template./bigj-ac-40s.modeling.py Only_10_7 cross-links.csv all 4.0 move_ac 0.01 Triple(on a single processor; prependmpirun -np 4or similar if you built IMP with MPI support)
The calculation will generate a trajectory, output/rmfs/0.rmf3, in
RMF format. For convenience, the 100
best-scoring models are also output in PDB format in the output/pdbs
directory.
Author(s): Riccardo Pellarin, Peter Cimermančič
License: LGPL. This library is free software; you can redistribute it and/or modify it under the terms of the GNU Lesser General Public License as published by the Free Software Foundation; either version 2 of the License, or (at your option) any later version.
Testable: Yes.
Parallelizeable: Yes
Publications:
- J. Erzberger, F. Stengel, R. Pellarin, S. Zhang, T. Schaefer, C. Aylett, P. Cimermančič, D. Boehringer, A. Sali, R. Aebersold, N. Ban. Molecular architecture of the 40S•eIF1•eIF3 translation initiation complex, Cell 158, 1125-1135, 2014.



 Download files
Download files Verified to work with the
Verified to work with the  To install the software needed to reproduce this system with the
To install the software needed to reproduce this system with the
 To set up the environment on the UCSF Wynton cluster to run
this system, run:
To set up the environment on the UCSF Wynton cluster to run
this system, run: