These scripts demonstrate the use of IMP, MODELLER, and PMI in the modeling of the proteasome/ecm29 complex using DSSO chemical cross-links.
Representation of ecm29 relied on (i) comparative models of 2 ecm29 domains built with MODELLER 9.17 (Sali and Blundell, 1993) based on the known related structures detected by HHPred (Soding, 2005; Soding et al. 2005) and (ii) secondary structure and disordered regions predicted by PSIPRED based on the ecm29 sequence (Buchan et al., 2013; Jones, 1999); see file ecm29.hhp in the comparative_modeling directory.
The sequences of the two templates (1U6G and 3W3W) and ecm29 can be found in template.ali.
The two templates have PDB code 1U6G and 3W3W respectively, with alignment to ecm29 in aligs.pir.
To obtain the comparative models, run model_ecm29_352.py and
model_ecm29_686.py.
63 DSSO cross-links involving ecm29 were identified via mass spectrometry; 56 of these cross-links were intramolecular and seven were intermolecular with the 19S proteasome, informing respectively the ecm29 conformation and the localization of ecm29 relative to the proteasome.
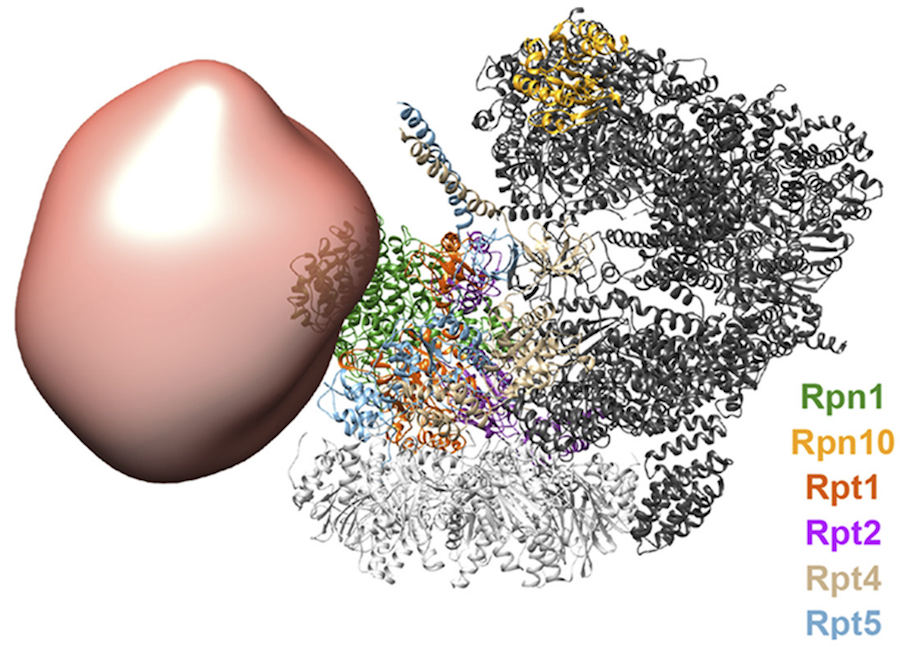
The 26S proteasome structure used was obtained from the PDB (code 5GJR); it was determined primarily based on a cryo-EM density map at 3.8Å resolution (EMDB code: 9508) {Huang, 2016 NSMB}.
-
smodeling.py: PMI modeling scripts for running the production simulations: The search for good-scoring models relied on Gibbs sampling, based on the Metropolis Monte Carlo algorithm. We suggest producing at least 3,750,000 models from 500 independent runs, each starting from a different initial conformation of ecm29 to have proper statistics. -
job_26s.sub: SGE cluster based submission script to run automatically 500 independent runs. -
The compressed 500 independent trajectories are accessible at:
/salilab/park1/ilan/ECM29/ECM29_19S/modeling[0-499].tar
Various scripts to analysis the simulations. we give more details scripts that allows us to test for sampling convergence.
-
Select_Top_Scoring_models.sh: basic bash scripts that read and summarize information found in the stat file generated by PMI. -
Random_Subset.sh: we test if adding more models improves our sampling of top scores. The input to the script is a list of all theTotal_Scorefrom the simulations. For each subset, we perform 100 sub-samplings to compute error bars. We give the file twice to check how the error bars vary. -
MannWhitney.py: we test if the distribution from two independent subsets are not unsimilar. -
The good-scoring models that have been selected for precision-based clustering based on RMSD metric are located in:
/salilab/park1/ilan/ECM29/ECM29_19S/Clustered_Models.tar.gzThe file name format is${trajectory_number}_${frame_number}.rmf3 -
Precision_Pvalue_Calculation.py: Given an RMSD matrix, we compute the 𝛘2-test for homogeneity of proportions{McDonald, 2014} and compute the best precision for which sampling has converged. -
RMSF_ECM29.py: Given a list of structures, compute the average RMSF, which indicates the precision of the structures in a cluster.
GNUPlot scripts for generating figures from the paper.
Author(s): Ilan E. Chemmama
Date: November 3rd, 2016; Updated August 16, 2017.
License: CC BY-SA 4.0 This work is licensed under the Creative Commons Attribution-ShareAlike 4.0 International License.
Testable: Yes
Parallelizeable: Yes
Publications: Xiaorong Wang, Ilan E Chemmama, Clinton Yu, Alexander Huszagh, Yue Xu, Rosa Viner, Sarah Ashley Block, Peter Cimermancic, Scott D Rychnovsky, Yihong Ye, Andrej Sali, and Lan Huang The Proteasome-Interacting Ecm29 Protein Disassembles the 26S Proteasome in Response to Oxidative Stress J. Biol. Chem. jbc.M117.803619. doi:10.1074/jbc.M117.803619



 Download files
Download files Verified to work with the
Verified to work with the  To install the software needed to reproduce this system with the
To install the software needed to reproduce this system with the
 To set up the environment on the UCSF Wynton cluster to run
this system, run:
To set up the environment on the UCSF Wynton cluster to run
this system, run: