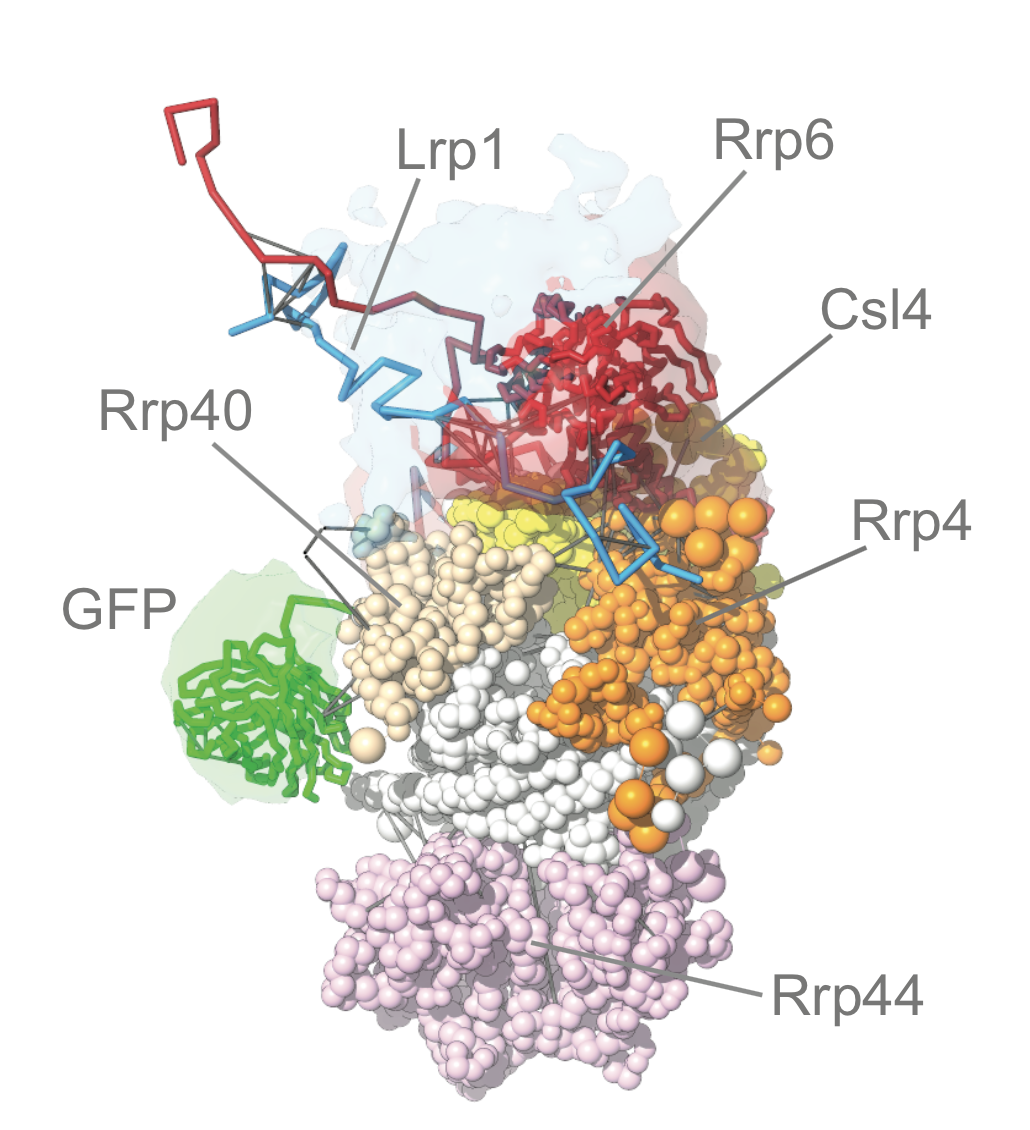
This repository contains the modeling scripts, the ensemble of models, and the results of the analysis for the modeling of the S.cerevisiae exosome complex (named exo10, comprising of Rrp40, Rrp4, Csl4, Rrp45, Rrp46, Rrp42, Rrp43, Mtr3, Ski6 and Rrp44), in presence of Ski7 (exo10+Ski7 below) or Rrp6 (exo10+Rrp6 below) proteins. The modeling is performed based on cross-link Mass-Spectrometry data, crystallographic structures, and comparative models.
To display the models and the localization densities using UCSF Chimera access ./Rrp6.analysis/kmeans_weight_500_2/cluster.1/ or ./Ski7.analysis/kmeans_weight_500_2/cluster.1/ and run chimera chimera.session.py
For more details on how to install IMP, run the modeling scripts and analyse the results using IMP and IMP.pmi see the IMP tutorial.
To run the modeling script, access: sampling/modeling and then run with:
python modeling.py
We recommend building IMP with MPI support, then running the modeling script with 64 replicas by prepending mpirun -np 64 to the above command.
modeling-scripts_Rrp6.1 modeling scripts and output modeling data for exo10+Rrp6 complex (first calculation)
modeling-scripts_Rrp6.2 modeling scripts and output modeling data for exo10+Rrp6 complex (second calculation)
modeling-scripts_Ski7.1 modeling scripts and output modeling data for exo10+Ski7 complex (first calculation)
modeling-scripts_Ski7.2 modeling scripts and output modeling data for exo10+Ski7 complex (second calculation)
README.md this readme file
Rrp6.analysis analysis for exo10+Rrp6 complex
Ski7.analysis analysis for exo10+Ski7 complex
data cross-linking data, fasta files, pdbs needed for the calculation
metadata files needed for the repository
test files needed to run tests
exosome.modeling.py modeling script (see above)
job.sh computer-cluster submission script (for UCSF QB3 cluster)
output all output data
output/pdbs best scoring 500 pdb files
output/rmfs rmf files for each replica
output/stat.*.out stat files containing modeling data (the index is the index of replica). To be read
using the python program IMP_source/modules/pmi/pyext/src/process_output.py
output/stat_replica.*.out stat files containing replica exchange data. To be read using IMP_source/modules/pmi/pyext/src/process_output.py
clustering.py: clustering script
kmeans_weight_500_2: the set of 500 best scoring models collected in two clusters
precision_rmsf.py: calculate the precision of clusters, their mutual distance and the RMSF
XL_table.py: calculate the contact map and the cross-link map
cluster.0: data for cluster 1
cluster.1: data for cluster 2
precision.0.0.out,precision.0.1.out,... : files containing the precision of a cluster (i.e., the files with the same indexes, precision.i.i.out, e.g., precision.0.0.out) and the files containing the distance between the clusters (i.e., files with different indexes, precision.i.j.out).
0.pdb,1.pdb,2.pdb....: the pdb files of the solutions
0.rmf3,1.rmf3,2.rmf3,...: the rmf files of the solution (can be opened with UCSF Chimera)
rmsf.Ski7.dat,rmsf.Rrp6.dat,...: text file of the RMSF analysis
rmsf.Ski7.pdf,rmsf.Rrp6.pdf,...: pdf file of the RMSF analysis
chimera_session.py: Chimera session file to display the localization densities
stat.out: stat file containing all relevant information on the score, etc.
XL_table_tail.pdf: pdf file of the cross-link map for the complex
License: LGPL. This library is free software; you can redistribute it and/or modify it under the terms of the GNU Lesser General Public License as published by the Free Software Foundation; either version 2 of the License, or (at your option) any later version.
Testable: Yes.
Parallelizeable: Yes
Publications:
- Y. Shi, R. Pellarin, P. Fridy, J. Fernandez-Martinez, M. Thompson, Y. Li, Q.J. Wang, A. Sali, M. Rout, B. Chait. A strategy for dissecting the architectures of native macromolecular assemblies, Nat Methods 12, 1135-8, 2015.



 Download files
Download files Verified to work with the
Verified to work with the  To install the software needed to reproduce this system with the
To install the software needed to reproduce this system with the
 To set up the environment on the UCSF Wynton cluster to run
this system, run:
To set up the environment on the UCSF Wynton cluster to run
this system, run: