news description
download install
examples manual
references
 |
ModView
Graphical user interface and Database interface to sequence
alignments and 3D structures of biomolecules.
by Valentin
ILYIN and Andrej SALI.
Laboratories
of Molecular Biophysics
Pels
Family Center for Biochemistry and Structural Biology
The
Rockefeller University
1230
York Avenue, New York, NY 10021-6399, USA
Copyright 2000, 2001 by Valentin Ilyin and Andrej Sali.
Modview was written at The Rockefeller University.
Any selling of the original or a modified program or its parts
is prohibited without a written permission from the authors.
|
|
 |
page under construction, please be patient
new page
News
-
Sep 7, 2001.
version 0.892 of ModView, (see Download) .
New commands
plugin: show ali
plugin: hide ali
plugin: show sliders
plugin: hide sliders
These commands can be used from command line and also at the initial call
(for example: script="plugin: hide ali"),
if you want to have specific combination of windows at first call of ModView.
I have also included checking between a sequence from the alignment and
corresponding PDB sequence. (they should be 100% identical)
There are several new input/output options in 'File' menu. you can load
alignment from file from html link, save ali to file, save
picture of the alignment as Postscript file. Load PDB coordinates
by PDB code.
Sort menu is added. you can sort sequences by ID, by Group (after
creating groups with tree), by length, by tree order.
-
July 23, 2001. version
ModView 0.889 for SGI (see Download)
For SGI it is better to copy both plugin files to the
root Netscape directory, which may looks like
/usr/local/netscape/plugins/
This may require some help from your SysAdm (at least the root password).
-
May 27, 2001. version
ModView 0.879 (see Download),
Many thanks to everybody for comments
!!!
current updates
-
fit by alignment works correctly for multiple proteins,
-
colors work properly for 'crazy' proteins (with non-sequencial
residue numbering),
-
you also may use different color scheme in ali-window,
-
I include checking on correspondence of sequence from alignment
and sequence from pdb structure,
-
error message window.
-
April 27, 2001. version
ModView 0.875, home page created.
Description
ModView is a program to visualize and
analyze multiple biomolecule structures and/or sequence alignments. As
a Netscape plugin, like
Chime
,
it can be embed into Web pages and controlled by Javascript objects on
the page.
Currently ModView is also used as Database
interface in several structure-sequence protein resources.
-
ModBase,
a Database of Comparative Protein Structure Models,
-
LigBase,
a Structural Database of Aligned Ligand Binding Protein Sequences,
-
DBAli,
a Database of Sequence-Structure alignments.
-
ModView can be also used to view structures and sequences
in
Protein Data Bank ,
choose 'Rasmol' display option on View Structure window.
Highlights
-
multiple structures representation and manipulation (as
many as you have memory).
-
fast manipulation with structures
-
manually or automatically edit alignments.
-
automatic superposition of biomolecules providing sequence-based
structural alignment.
-
extended coloring and viewing options.
-
quality printing output.
-
internet or local connection to structural and/or alignment
databases.
Download.
Installation.
Needs: ModView contains two parts, C++ and Java
part, both parts are necessary for full functionality of the program. To
install ModView just copy these two files to your ~/.netscape/plugins directory.
How to do it step by step.
-
Create directory ~/.netscape/plugins
. In case you do not have it.
-
Check how many colors do you have
in your computer. 32,24 or 16 bit graphical card. Current desktops usually
have 32 bit cards and laptops have 16 bit.
-
Download two files. File 1 (C++ part)
is specific for your display. For Linux 6.2+ with 32 or 24 bit graphical
card use npmodview-32-XXXX.so , or
for 16 bit graphical card use npmodview-16-XXXX.so.
Only one file 1 should be used. Second file is common, use modview-XXXX.zip,
do not unzip it!
-
Copy these two files in your ~/.netscape/plugins
directory and restart Netscape.
-
Set preferences in Netscape window.
Go to Edit-> Preferences-> Navigator-> Applications and set next Suffixes
for corresponding MIME types. For most users only two Suffixes should be
set : pdb for 'chemical/x-pdb' and ali for 'protein/x-ali'.
More experienced users may check all the Suffixes for all supported MIME
types by ModView in Help->About Plugins.
-
To check how it works, there are
some examples.
Snapshots
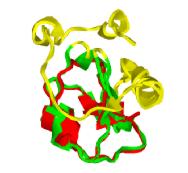
123 crystal structures and amino-acid sequences of
myoglobins and hemoglobins
other snapshots
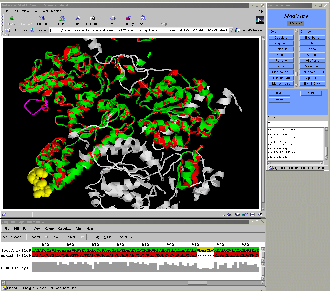
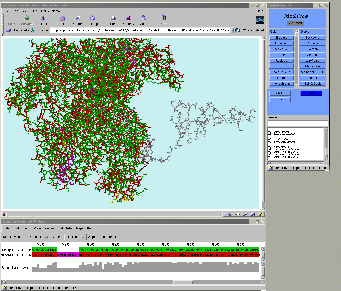
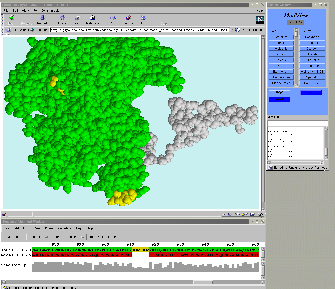
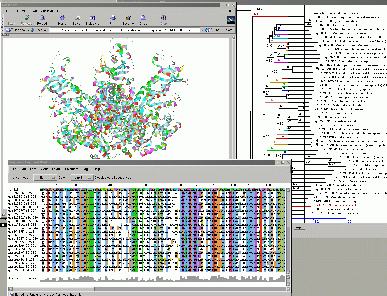
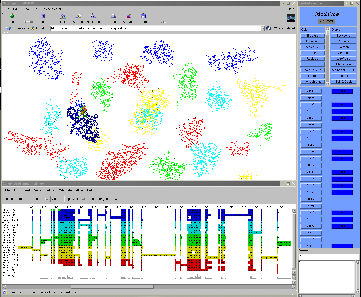
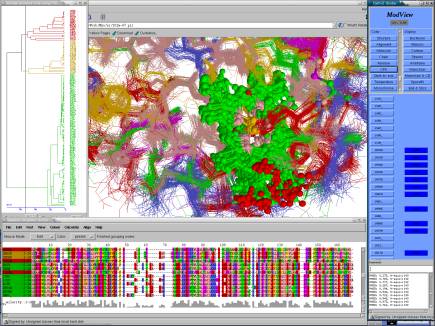
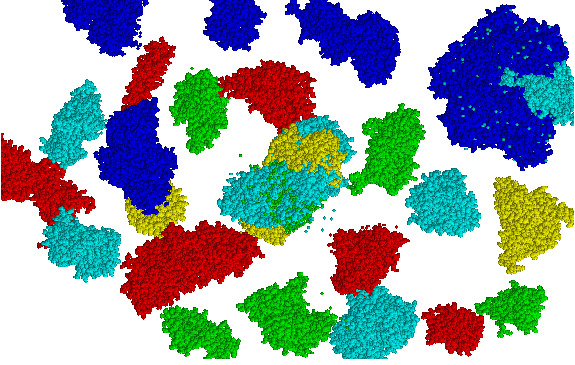 26 proteins, more than 100,000 atoms interactively!!!
26 proteins, more than 100,000 atoms interactively!!!
Examples
To see examples the plugin should be already installed, see Download
and Installation.
-
(1D2R)
,
plain PDB file Ligand-Free Tryptophanyl-tRNA Synthetase by Valentin
A. Ilyin and Charles W. Carter Jr.
-
(5FD1)
and (1FDX) , two proteins alignment Azotobaster vinelandii ferredoxin
by C.D. Stout and Peptococcus aerogen ferredoxin by E.T.Adman et.
al.
-
(Many)
, seven proteins of T1 ribonuclease family.
User
Manual help is coming ... help
The user interface is easy-to-use, all menus and
buttons have 'common protein sense' ;-) . To raise menu for structures
place mouse on structure window and press right button (do not move mouse,
if move the structure moves). On control panel (3d window with many buttons),
the middle vertical scroll bar is slab. There is input command field to
write Rasmol commands, and under it log text area. Fit functions works
good for two proteins, may crash for multiple structures.
References
ModView uses Roger Sayle's
Rasmol
rendering
system and accept Rasmol scripts.
ModView uses modified alignment editor panel from Michele Clamp's
Jalview
.
I would really appreciate your comments and suggestions, especially
bug reports.
Valya Ilyin. ilyin@salilab.org


 26 proteins, more than 100,000 atoms interactively!!!
26 proteins, more than 100,000 atoms interactively!!!