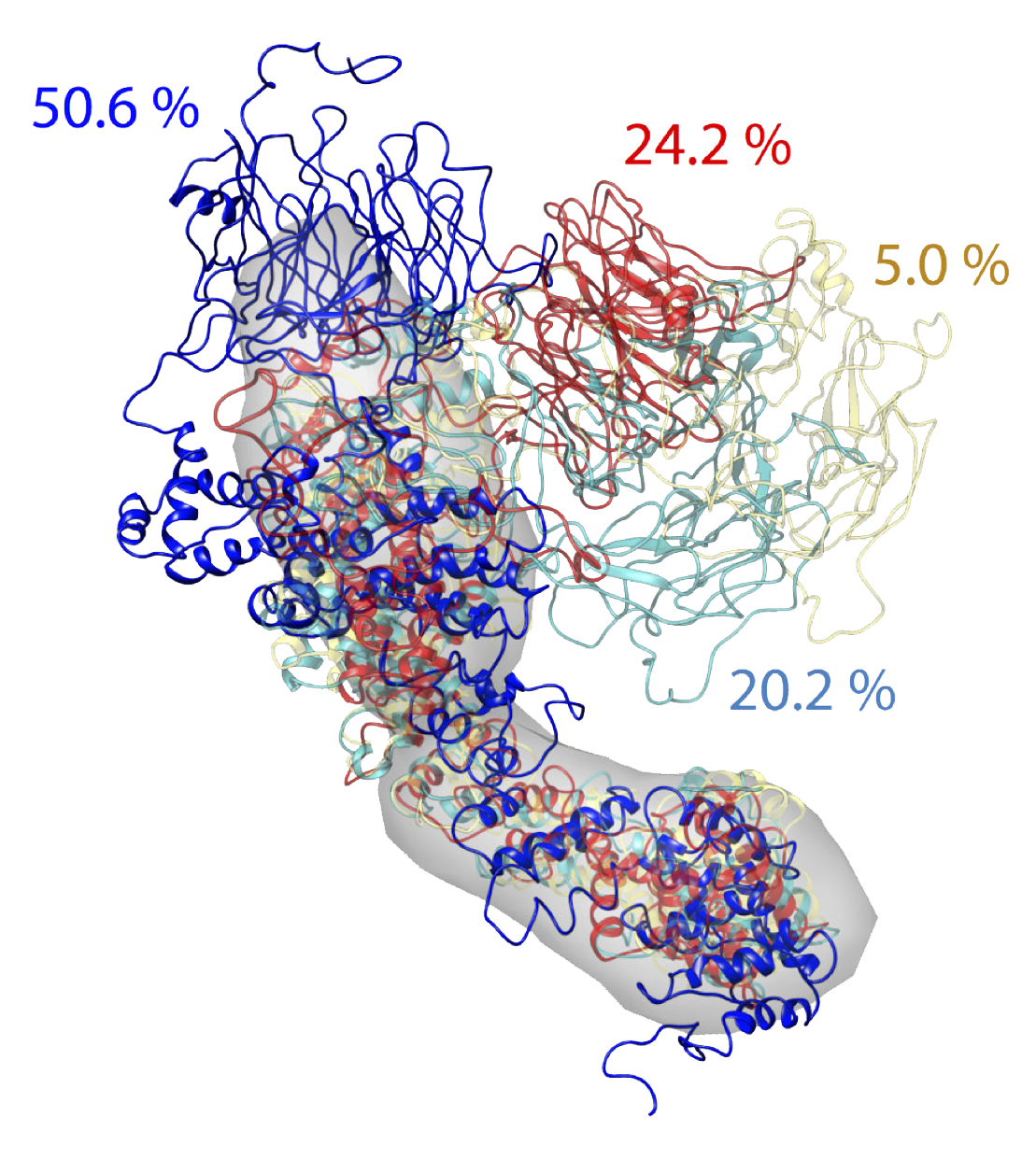
These scripts demonstrate the use of IMP, MODELLER, FoXS, and AllosMod, and Minimal Ensemble Search in the modeling of the Nup133 protein in the S. cerevisiae Nuclear Pore Complex (NPC). First, MODELLER is used to generate an initial comparative model of Nup133 guided by FoXS fits to SAXS data. Then, the model was subjected to conformational sampling with AllosMod, and finally Minimal Ensemble Search was used to identify four models that together reproduced both the SAXS data and a set of electron microscopy class averages. The final model was also validated against a set of chemical cross-links, that were not used in the modeling.
A full description of the scripts can be found in Integrative structure-function mapping of the nucleoporin Nup133 suggests a conserved mechanism for membrane anchoring of the nuclear pore complex.
Python scripts to generate comparative models of parts of the Nup133 can
be found in the MODELLER subdirectory. Each subdirectory corresponds to
modeling consistent with a single construct (directory names match the protein
IDs in Supplementary Table S1 in the publication, also available as
SAXS/Nup133_tableS1.xls). A number of candidate models were generated
for each construct, ranked by FoXS score, and the single best model selected
for subsequent steps.
all_sjkim_final.pyinMODELLER/23917generates models consistent with construct 490-1157all_sjkim_final_23902a.pyinMODELLER/23902generates models consistent with construct 2-515all_sjkim_final_23904.pyinMODELLER/23904Ctakes the best scoring model 23917 and generates models consistent with construct 490-1157all_sjkim_final_23904.pyinMODELLER/23904takes the best scoring model 23904C and 23902a and generates models consistent with construct 2-1157 (the complete protein)
The complete ensemble was generated by submission to the AllosMod web server.
Results from running Minimal Ensemble Search can be found in the
outputs_foxs_ensemble_new/MES_results directory. new_mes4.log contains
the final selected ensemble of four models, also present in this directory.
The electron microscopy class averages used are also present in the
outputs_foxs_ensemble_new/ISAC_p150_t346_m30 directory.
Intramolecular cross-link data is available in
Crosslinks/DSS_EDC_crosslinks.txt. Simple Python scripts to calculate
residue-residue distances for the final ensemble can be found in the same
directory.
Author(s): Seung Joong Kim
Date: September 2nd, 2014
License: CC BY-SA 4.0 This work is licensed under the Creative Commons Attribution-ShareAlike 4.0 International License.
Testable: Yes.
Parallelizeable: No
Publications:
- Seung Joong Kim*, Javier Fernandez-Martinez*, Parthasarathy Sampathkumar*, Anne Martel, Tsutomu Matsui, Hiro Tsuruta, Thomas Weiss, Yi Shi, Ane Markina-Inarrairaegui, Jeffery B. Bonanno, J. Michael Sauder, Stephen K. Burley, Brian T. Chait, Steven C. Almo, Michael P. Rout, and Andrej Sali, Integrative structure-function mapping of the nucleoporin Nup133 suggests a conserved mechanism for membrane anchoring of the nuclear pore complex, Molecular & Cellular Proteomics, 2014, mcp.M114.040915.
*These authors contributed equally to this work as co-first authors.



 Download files
Download files Verified to work with the
Verified to work with the  To install the software needed to reproduce this system with the
To install the software needed to reproduce this system with the
 To set up the environment on the UCSF Wynton cluster to run
this system, run:
To set up the environment on the UCSF Wynton cluster to run
this system, run: