These scripts demonstrate the use of IMP, MODELLER and Chimera in the modeling of the phosphodiesterase (PDE6).
MODELLER is used to generate initial models for the structure using multiple templates
(PDB codes 1tbf, 3bjc, 3dba, 3ibj). Cross links, symmetry and secondary structure restraints are used in modeling:
cd comparative_modelling; ./model_mult.py
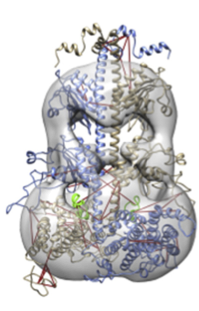
The initial models do not fit well into the 3D EM density map (20Å resolution). Therefore, IMP is used to fit the complex into the electron microscopy density map.
Usage and content of the directory integrative_modeling
-
access the directory:
cd integrative_modeling -
test the python script:
./run_modeling.py testno error = all tests passed -
run sampling:
./run_modeling.pyall output data will be stored in output/ -
analyse the results:
3.1) compile the clustering algorithm in bin/
gfortran cluster.f u3best.f -o cluster.x
3.2) run the analysis script:
bin/get_frames.sh
100 best scoring frames will be stored in best_pdb/
and clustering data will be stored in clustering/
Finally the final models are rebuilt and refined with Modeller:
cd model_refinement/cluster1; ./model-single.py
Author(s): Riccardo Pellarin, Dina Schneidman
Version: 1.0
License: LGPL. This library is free software; you can redistribute it and/or modify it under the terms of the GNU Lesser General Public License as published by the Free Software Foundation; either version 2 of the License, or (at your option) any later version.
Testable: Yes.
Parallelizeable: No
Publications:
- X. Zeng-Elmore, X. Gao, R. Pellarin, D. Schneidman-Duhovny, X. Zhang, K. Kozacka, Y. Tang, A. Sali, R. Chalkley, R. Cote, F. Chu. Molecular architecture of photoreceptor phosphodiesterase elucidated by chemical cross-linking and integrative modeling, J Mol Biol 426, 3713-3728, 2014.



 Download files
Download files Verified to work with the
Verified to work with the  To install the software needed to reproduce this system with the
To install the software needed to reproduce this system with the
 To set up the environment on the UCSF Wynton cluster to run
this system, run:
To set up the environment on the UCSF Wynton cluster to run
this system, run: