The 'PAP' format, which corresponds to a relatively nice looking alignment, has several additional formatting options that can be selected by the ALIGNMENT_FEATURES variable. This scalar variable can contain any combination of the following keywords:
- 'INDICES', the alignment position indices;
- 'CONSERVATION', a star for each absolutely conserved position;
- 'ACCURACY', the alignment accuracy indices, scaled between 0-9,
as calculated by ALIGN_CONSENSUS;
- 'HELIX', average content of helical residues for
structures 1 - ALIGN_BLOCK at each position,
0 for 0% and 9 for 100%, as calculated by ALIGN2D.
- 'BETA', average content of
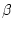 -strand residues for
structures 1 - ALIGN_BLOCK at each position,
0 for 0% and 9 for 100%, as calculated `by ALIGN2D.
-strand residues for
structures 1 - ALIGN_BLOCK at each position,
0 for 0% and 9 for 100%, as calculated `by ALIGN2D.
- 'ACCESSIBILITY', average relative sidechain buriedness for
structures 1 - ALIGN_BLOCK, 0 for 0% (100% accessibility) and
9 for 100% (0% accessibility), as calculated by ALIGN2D;
- 'STRAIGHTNESS', average mainchain straightness structures 1 - ALIGN_BLOCK at each position 0 for 0% and 9 for 100%, as calculated by ALIGN2D.
Options 'HELIX', 'BETA', 'ACCESSIBILITY', and 'STRAIGHTNESS' are valid only after executing command ALIGN2D, where the corresponding quantities are defined. They refer to the 3D profile defined for the first ALIGN_BLOCK structures (run ALIGN2D with FIT = off to prepare these structural data without changing the input alignment). Similarly, the 'ACCURACY' option is valid only after the CONSENSUS_ALIGNMENT command.
ALIGN_ALIGNMENT and ALIGN_BLOCK are used to ensure correct indication of identical alignment positions, depending on whether sequences or two blocks of sequences were aligned: For sequences (ALIGN_ALIGNMENT = off and ALIGN_BLOCK is ignored), a '*' indicating a conserved position is printed where all sequences have the same residue type. For blocks (ALIGN_ALIGNMENT = on and ALIGN_BLOCK indicates the last sequence of the first block), a '*' is printed only where the two blocks have the same order of residue types (there has to be the same number of sequences in both blocks). The blocks option is useful when comparing two alignments, possibly aligned by the ALIGN command.