

|
TutorialDifficult example:
|
| Conf. | Net Score | E-value | PairE | SolvE | Aln Score | Aln Len | Str Len | Seq Len | Alignment | SCOP Codes |
|---|---|---|---|---|---|---|---|---|---|---|
| CERT | 0.903 | 1e-04 | -516.4 | -0.7 | 232.0 | 166 | 180 | 298 | 1ej0A0 | c.66.1.2 |
| MEDIUM | 0.650 | 0.02 | -512.7 | 1.7 | 114.0 | 151 | 173 | 298 | 1j4fA0 | - |
| MEDIUM | 0.645 | 0.022 | -502.6 | -2.7 | 122.0 | 155 | 230 | 298 | 1fbnA0 | c.66.1.3 |
| MEDIUM | 0.640 | 0.024 | -467.5 | -3.9 | 121.0 | 152 | 194 | 298 | 1dusA0 | c.66.1.4 |
| MEDIUM | 0.620 | 0.038 | -435.7 | -2.6 | 120.0 | 159 | 264 | 298 | 1i9gA0 | c.66.1.13 |
| MEDIUM | 0.606 | 0.05 | -485.2 | -1.6 | 115.0 | 166 | 186 | 298 | 1kxzA0 | c.66.1.22 |
Extracts from mGenThreader results. File: 30133975_mGenThreader.html
Alignment between the nsp16 sequence and the 1ej0A from mGenThreader results.
C; mGenThreader alignment of 30133975 and 1ej0A
C; CERT significance eith an e-value of 1e-04
C; Percentage Identity = 14.4%
>P1;1ej0A
structureX:1ej0: 30 :A: 209 :A::::
-------GLRSRAWFKL----------------------------------DEIQQSDKLFKPGMTVVDL
GA------APGGWSQYVVTQIGGKGRIIACDLLPMDPIVGVDFLQGDFRDELVMKALLERVGDSKVQVVM
SDMAPNMSGTPAVDIPRAMYLVELALEMCRDVLAPGGSFVVKVFQGEGFDEYLREIRSLFTKVKVRKPDS
SRARSREVYIVATGRKP*
>P1;30133975
sequence:::::::::
ASQAWQPGVAMPNLYKMQRMLLEKCDLQNYGENAVIPKGIMMNVAKYTQLCQYLNTLTLAVPYNMRVIHF
GAGSDKGVAPG--TAVLRQWLPTGTLLVDSDLNDFVSDADSTLIGDCATVH----------TANKWDLII
SDMYDPRTKHVTKENDSKEGFFTYLCGFIKQKLALGGSIAVKITEHS-WNADLYKLMGHFSWWTAFVTNV
NA-SSSEAFLIGANYLG*
File 30133975_1ej0A_mGenThreader.ali. Red residues were manually removed from the alignment.
Five models were built for the nsp16 sequence based on the mGenThreader alignment. The file model.top shows the TOP script used.
INCLUDE SET ALNFILE = '30133975_1ej0A_mGenThreader.ali' SET KNOWNS = '1ej0A' SET SEQUENCE = '30133975' SET STARTING_MODEL = 1 SET ENDING_MODEL = 5 CALL ROUTINE = 'model'
File: model.top
All 5 models were then evaluated with the program PROSAII and the model 30133975.B99990001 was selected as the final model.
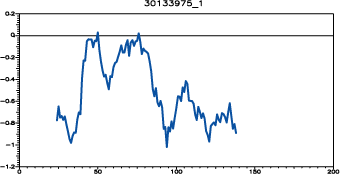
PROSAII model evaluation for 30133975_1 model
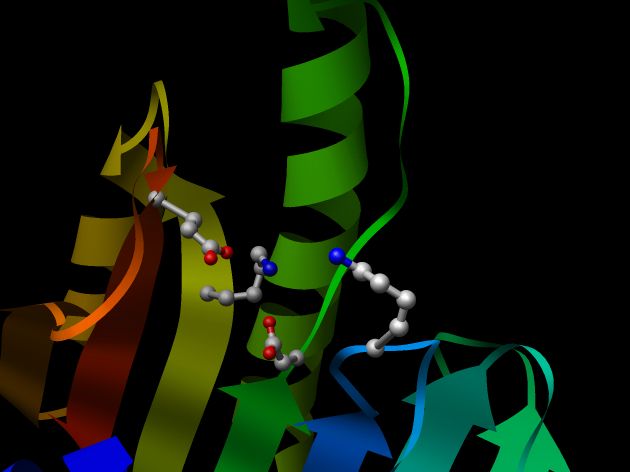
Figure of the model 30133975_1 rendered with RasMol
The PDB structure 1ej0A corresponds to a mRNA cap methylation. These proteins are found indispensable for efficient replication of many viruses and represents an active area for drug development. Nevertheless, direct inhibitors of the nsp13 enzyme may fail to suppress viral replication, as the cap-1 formation seems to be less critical than the preceding cap-0 (mGpppN) formation. The existence of the cap-1-forming enzyme in the genome would suggest that the virus also requires the AdoMet-dependent cap-0 methyltransferase. Both functions can be inhibited by carbocyclic analogs of adenosine, such as Neplanocin A or 3-deazaneplanocin A, which interfere with the AdoMet-AdoHcy metabolism of the host cell. Those compounds could complement other therapeutic strategies aimed at blocking enzymatic functions such as the RNA-dependent RNA polymerase, the protease, or the helicase encoded by the SARS virus.
This exercise was inspired by the work of Grotthuss, Wyrwicz and
Rychlewski
Letter to the Editor
"mRNA Cap-1 Methyltransferase in the SARS Genome"
Marcin von Grotthuss, Lucjan S. Wyrwicz, and Leszek Rychlewski Cell, Vol 113,
701-702, 13 June 2003
 |
MODELLER (copyright © 1989-2004 Andrej Sali) is maintained by Ben Webb at the Departments of Biopharmaceutical Sciences and Pharmaceutical Chemistry, and California Institute for Quantitative Biomedical Research, Mission Bay Genentech Hall, University of California San Francisco, San Francisco, CA 94143-2240, USA. Any selling or distribution of the program or its parts, original or modified, is prohibited without a written permission from Andrej Sali. This file last modified: Fri May 6 13:26:20 PDT 2005. |