Tutorial
Iterative example:
The alignment-modeling-evaluation cycle. The case of the
Holoferax volcanii dihydrofolate reductase.
All input and output files for this example are available to download,
in either zip format (for Windows) or
.tar.gz format (for Unix/Linux).
Several structures of dihydrofolate reductase (DHFR) are known. However,
the structure of DHFR from Haloferax volcanii was not known and its
sequence identity with DHFRs of known structure is rather low ~30%. A model
of H. volcanii DHFR (HVDFR) was constructed before the experimental
structure was solved. This example illustrates the power of the iterative
alignment-modeling-evaluation approach to comparative modeling.
Of all the available DHFR structures, HVDHFR has the sequence most
similar to DHFR from E. coli. The PDB entry
4DFR corresponds to a high resolution (1.7Å)
E. coli DHFR structure. It contains two copies of the molecule,
named chain A and chain B. According to the authors, the structure for chain
B is of better quality than that of chain A. The following TOP file aligns
HVDFR and chain B of 4DFR.
READ_MODEL FILE = '4dfr.pdb', MODEL_SEGMENT = 'FIRST:B' 'LAST:B'
SEQUENCE_TO_ALI ALIGN_CODES = '4dfr'
READ_ALIGNMENT FILE = 'hvdfr.seq', ALIGN_CODES = ALIGN_CODES 'hvdfr', ADD_SEQUENCE = on
ALIGN2D
WRITE_ALIGNMENT FILE='hvdfr-4dfr.ali'
WRITE_ALIGNMENT FILE='hvdfr-4dfr.pap', ALIGNMENT_FORMAT = 'PAP', ;
ALIGNMENT_FEATURES = 'indices helix beta'
File: align2d-4.top
Some options used in this example include MODEL_SEGMENT,
which is used to indicate chain B of 4DFR; and
ALIGNMENT_FEATURES, which is used to output information
such as secondary structure, to the alignment file in the PAP format.
_aln.pos 10 20 30 40 50 60
4dfr -MISLIAALAVDRVIGMENAMPW-NLPADLAWFKRNTLDKPVIMGRHTWESIGRPLPGRKNIILSSQP
hvdfr MELVSVAALAENRVIGRDGELPWPSIPADKKQYRSRIADDPVVLGRTTFESMRDDLPGSAQIVMSRSE
_helix 999999999999 999999999
_beta 9999999999 999999 99999999
_aln.p 70 80 90 100 110 120 130
4dfr GTDDRVTWVKSV----DEAIAACGDVPEIMVIGGGRVYEQFLPKAQKLYLTHIDAEVEGDTHFPDYEP
hvdfr RSFSVDTAHRAASVEEAVDIAASLDAETAYVIGGAAIYALFQPHLDRMVLSRVPGEYEGDTYYPEWDA
_helix 99 99999999 99999999
_beta 99999 9999999 999999999
_aln.pos 140 150 160
4dfr DDWESVFSEFHDADAQNSHSYCFKILERR
hvdfr AEWELDAETDHEG---FTLQEWVRSASSR
_helix
_beta 999999999999 999999999999
File: hvdfr-4dfr.pap
Using the PIR alignment file hvdfr-4dfr.ali,
an initial model is calculated.
INCLUDE
SET ALNFILE = 'hvdfr-4dfr.ali'
SET KNOWNS = '4dfr'
SET SEQUENCE = 'hvdfr'
SET STARTING_MODEL = 1
SET ENDING_MODEL = 1
CALL ROUTINE = 'model'
File: model4.top
Because the sequence identity between 4DFR and
HVDFR is relatively low (30%), the automated alignment is likely to contain
errors. The PROSA evaluation of the model shows two
regions with positive energy.
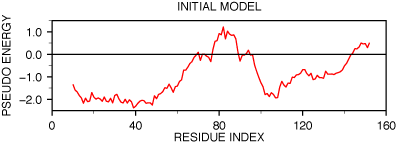
PROSAII profile for model initial model
The first region is around residue 85, the second region is at the
C-terminal end of the protein. Referring to the target--template alignment
above (hvdfr-4dfr.pap), it is easy to understand
why the first positive peak appears. The insertion between position 85 and
88 of the alignment was placed in the middle of an α-helix in the
template (the "9" characters on the first line below the sequence
mark the helices). Moving the insertion to the end of the α-helix may
improve the model.
The second problem, which occurs in the C-terminal region of the alignment,
is less clear. The deletion in that region of the alignment corresponds to the
loop between the last two β-strands of 4DFR
(a β-hairpin). Since the profile suggests that this region is in error,
an alternative alignment should be tried. One possibility is that the deletion
is actually longer, making the C-terminal β-hairpin shorter in HVDFR.
One plausible alignment based on these considerations is shown here.
_aln.pos 10 20 30 40 50 60
4dfr M-ISLIAALAVDRVIGMENAMPW-NLPADLAWFKRNTLDKPVIMGRHTWESIGRPLPGRK
hvdfr MELVSVAALAENRVIGRDGELPWPSIPADKKQYRSRIADDPVVLGRTTFESMRDDLPGSA
_helix 999999999999 999999999
_beta 9 999999999 999999 999
_aln.pos 70 80 90 100 110 120
4dfr NIILSSQPGT--DDRVTWVKSVDEAIAACG--DVPEIMVIGGGRVYEQFLPKAQKLYLTH
hvdfr QIVMSRSERSFSVDTAHRAASVEEAVDIAASLDAETAYVIGGAAIYALFQPHLDRMVLSR
_helix 9999999999 99999999
_beta 99999 99999 9999999 9999999
_aln.pos 130 140 150 160
4dfr IDAEVEGDTHFPDYEPDDWESVFSEFHDADAQNSHSYCFKILERR----
hvdfr VPGEYEGDTYYPEWDAAEWELDAETDHE-------GFTLQEWVRSASSR
_helix
_beta 99 999999999999 999999999999
File: hvdfr-4dfr-2.pap
A new model was calculated using this alignment and the TOP script,
modified to use the new alignment (see file
`model5.top'). Its
PROSA profile is shown in the next figure.
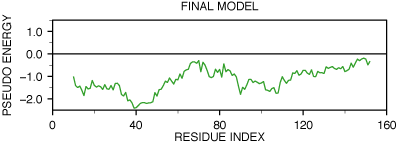
PROSAII profile for model final model
Both positive peaks disappeared and the new profile does not contain any
positive regions. Next figure shows the comparison of the C-terminal
beta-hairpin of both models and the actual experimental structure. This
confirms that the correct choice for the final alignment was made and that
PROSA was indeed able to detect the error in the
initial alignment.
| 

