



Next: alignment.get_suboptimals() parse
Up: The alignment class: comparison
Previous: alignment.malign3d() align
Contents
Index
Subsections
salign(residue_type2='REGULAR', no_ter=False, overhang=0, off_diagonal=100, matrix_offset=0.0, gap_penalties_1d=(-900.0, -50.0), gap_penalties_2d=(3.5, 3.5, 3.5, 0.2, 4.0, 6.5, 2.0, 0.0, 0.0), gap_penalties_3d=(0.0, 1.75), feature_weights=(1.0, 0.0, 0.0, 0.0, 0.0, 0.0), rms_cutoff=3.5, fit=True, surftyp=1, fit_on_first=False, gap_function=False, align_block=0, max_gap_length=999999, align_what='BLOCK', input_weights_file=None, output_weights_file=None, weigh_sequences=False, smooth_prof_weight=10, fix_offsets=(0.0, -1.0, -2.0, -3.0, -4.0), substitution=False, comparison_type='MAT', matrix_comparison='CC', alignment_type='PROGRESSIVE', edit_file_ext=('.pdb', '_fit.pdb'), weights_type='SIMILAR', similarity_flag=False, bkgrnd_prblty_file='$(LIB)/blosum62_bkgrnd.prob', ext_tree_file=None, dendrogram_file='', matrix_scaling_factor=0.0069, auto_overhang=False, overhang_factor=0.4, overhang_auto_limit=60, local_alignment=False, improve_alignment=True, fit_atoms='CA', output='', write_whole_pdb=True, current_directory=True, write_fit=False, fit_pdbnam=True, rr_file='$(LIB)/as1.sim.mat', n_subopt=0, subopt_offset=0.0, align3d_trf=False, normalize_pp_scores=False, gap_gap_score=0.0, gap_residue_score=0.0, nsegm=2, matrix_offset_3d=-0.1, io=None)
- Output:
- SalignData object
This command is a general dynamic programming based alignment
procedure for aligning sequences, structures or a combination of the two. It
is loosely based on the program COMPARER [Šali & Blundell, 1990]. SALIGN can be used
to generate multiple protein structures/sequences alignments or to align two
blocks of sequences/structures that are in memory.
See also Section 6.32 for utility scripts to simplify the
high-level usage of SALIGN.
Please note that the method is still in development, and has not yet been
fully benchmarked. As with any other alignment method, generated alignments
should be assessed for quality.
Broadly classifying, three different types of protein alignment categories
are tackled by this command:
- Multiple structure alignments
- Aligning a structure block to a sequence block
- Multiple and pair-wise protein sequence alignment
The command incorporates the functionality of several old MODELLER commands
(alignment.align(), alignment.align2d(), alignment.malign(),
alignment.align3d(), and alignment.malign3d()). Some of the examples
below illustrate the equivalent script files to replace the old alignment
commands with alignment.salign().
In addition to these, this command has several new alignment features
including profile-profile sequence alignments and a dendrogram based multiple
sequence/structure alignment among others.
All pair-wise alignments make use of local or global dynamic programming.
A switch from one to another can be effected by setting local_alignment
to True or False. The dynamic programming can be carried out using
affine gap penalties (as previously used in alignment.align(), by setting
gap_function to False) or an environment dependent gap penalty
function (as used in alignment.align2d(), by setting gap_function to
True). (Please note that the default gap_penalties_1d parameters
are optimal for the affine gap penalty; see the align2d examples for reasonable
parameters if you wish to use the environment dependent gap penalty.)
All arguments that associated to the alignment.align() and
alignment.align2d() commands apply.
If at least one of the blocks in a pairwise alignment consists of structures,
dynamic programming can be performed using structure dependent gap penalties.
On successful completion, an SalignData object is returned, from which
some of the calculated data can be queried. For example, if you save this in
a variable 'r', the following data are available:
- r.aln_score; the alignment score
- r.qscorepct; the quality score (percentage) if output contains
'QUALITY'
Central to the dynamic programming algorithm is the weight matrix. In SALIGN,
this matrix is constructed by weighting the contribution from six
features of protein structure and sequence:
- Feature 1
- is the residue type.
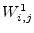 is obtained from the residue
type - residue type dissimilarity matrix, specified in the file rr_file.
is obtained from the residue
type - residue type dissimilarity matrix, specified in the file rr_file.
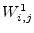 dissimilarity score for positions
dissimilarity score for positions 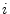 and
and 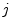 in the two compared
sub-alignments is the average dissimilarity score for a comparison of all
residues in one sub-alignment with all residues in the other sub-alignment
(note that gaps are ignored here). Should only feature weight 1 be non-zero,
the user has an option of considering residue-residue similarity scores
instead of distance scores by setting similarity_flag to True.
(The other features are distance features, and so if their weights are non-zero,
similarity_flag must be turned off, which is the default.)
in the two compared
sub-alignments is the average dissimilarity score for a comparison of all
residues in one sub-alignment with all residues in the other sub-alignment
(note that gaps are ignored here). Should only feature weight 1 be non-zero,
the user has an option of considering residue-residue similarity scores
instead of distance scores by setting similarity_flag to True.
(The other features are distance features, and so if their weights are non-zero,
similarity_flag must be turned off, which is the default.)
- Feature 2
- is the inter-molecular distance for a pair of residues (unless
align3d_trf is True: see alignment.align3d()). Only one atom per
residue is of course selected, as specified by fit_atoms (e.g.,
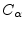 ,
although we should also allow for
,
although we should also allow for 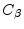 in the future, which requires an
intervention for Gly). This `position' feature is complicated because it
depends on the relative orientation of the structures corresponding to the two
compared alignments.
in the future, which requires an
intervention for Gly). This `position' feature is complicated because it
depends on the relative orientation of the structures corresponding to the two
compared alignments. 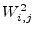 is the Euclidean distance between the
compared positions
is the Euclidean distance between the
compared positions 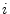 and
and 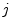 in the two compared sub-alignments that are
already optimally aligned and superposed based on their coordinates alone.
This optimal alignment is obtained by an iterative procedure as follows
(the same as in alignment.align3d()). The average structures for both
sub-alignments are calculated for all sub-alignment positions with at least
one defined selected atom. This calculation is straightforward because the
structures within the two sub-alignments are already superposed with each
other (see below). Then, the distance matrix for dynamic programming with affine
gap penalties is calculated as the matrix of Euclidean distances between
the two averages. The dynamic programming results into a new alignment,
dependent also on the gap initiation and extension penalties
gap_penalties_3d (a reasonable setting is (0, 3)).
gap_penalties_3d[0] is a gap creation penalty (usually 0), and
gap_penalties_3d[1] is a gap extension penalty, say 3. When the gap
initiation penalty is 0, pairs of positions are identified as equivalent
when they have their selected atoms at most 2 times gap_penalties_3d[1]
angstroms apart in the current superposition, as described for the
alignment.align3d() command. The new alignment is then used to generate
the new superposition of the two averages, and the iteration of the distance
matrix calculation, alignment and superposition is repeated until there are
no changes in the number of equivalent positions and in the rotation matrix
relating the two averages.
in the two compared sub-alignments that are
already optimally aligned and superposed based on their coordinates alone.
This optimal alignment is obtained by an iterative procedure as follows
(the same as in alignment.align3d()). The average structures for both
sub-alignments are calculated for all sub-alignment positions with at least
one defined selected atom. This calculation is straightforward because the
structures within the two sub-alignments are already superposed with each
other (see below). Then, the distance matrix for dynamic programming with affine
gap penalties is calculated as the matrix of Euclidean distances between
the two averages. The dynamic programming results into a new alignment,
dependent also on the gap initiation and extension penalties
gap_penalties_3d (a reasonable setting is (0, 3)).
gap_penalties_3d[0] is a gap creation penalty (usually 0), and
gap_penalties_3d[1] is a gap extension penalty, say 3. When the gap
initiation penalty is 0, pairs of positions are identified as equivalent
when they have their selected atoms at most 2 times gap_penalties_3d[1]
angstroms apart in the current superposition, as described for the
alignment.align3d() command. The new alignment is then used to generate
the new superposition of the two averages, and the iteration of the distance
matrix calculation, alignment and superposition is repeated until there are
no changes in the number of equivalent positions and in the rotation matrix
relating the two averages.
The values of both improve_alignment and fit are used in the
calculation of the position feature. That is, the initial alignment and
the orientation of the coordinates can be selected not to change at all
during the calculation of the inter-molecular distance matrix.
When the calculation of the inter-molecular distance matrix is finished, all
the structures in the second sub-alignment are rotated and translated
following the optimal rotation and translation of the second average on the
first average. These superpositions prepare the individual structures for the
next of the 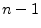 stages of the progressive multiple alignment, and also
orient all the structures for writing out to atom files with a '_fit.pdb'
extension if write_fit = True. If fit_pdbnam = False, the
PDB filenames in the output alignment file will not have the '_fit.pdb'
extensions. Thus, feature 2 needs to be selected by
feature_weight[2]
stages of the progressive multiple alignment, and also
orient all the structures for writing out to atom files with a '_fit.pdb'
extension if write_fit = True. If fit_pdbnam = False, the
PDB filenames in the output alignment file will not have the '_fit.pdb'
extensions. Thus, feature 2 needs to be selected by
feature_weight[2] 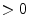 if you wish to write out the structures
superposed according to the tree-following procedure; also, fit_on_first
must be False, otherwise the structures are written out superposed on the
first structure according to the final alignment (see also below).
if you wish to write out the structures
superposed according to the tree-following procedure; also, fit_on_first
must be False, otherwise the structures are written out superposed on the
first structure according to the final alignment (see also below).
The alignment produced within the routine that calculates 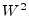 does not
generally correspond to the alignment calculated based on
does not
generally correspond to the alignment calculated based on 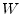 . Therefore, the
multiply superposed structures are not necessarily superposed based on the
final multiple alignment produced by alignment.salign(). If you wish such
a superposition, you can use alignment.malign3d() with fit = False
and write_fit = True (the meaning of fit is different between
alignment.salign() and alignment.malign3d()).
. Therefore, the
multiply superposed structures are not necessarily superposed based on the
final multiple alignment produced by alignment.salign(). If you wish such
a superposition, you can use alignment.malign3d() with fit = False
and write_fit = True (the meaning of fit is different between
alignment.salign() and alignment.malign3d()).
Unless the position feature is selected, the initial alignment does not matter.
If the position feature is selected, a good starting alignment is a multiple
sequence alignment, obtained either by alignment.malign() or by
alignment.salign() used without the position feature (the initial
alignment can also be prepared using the position feature). If the position
feature is used, each pair of structures needs to have at least 3 aligned
residues at all points during the alignment.
There are several possibilities as to the final orientation of the input
coordinates. If fit_on_first is True, all the coordinate sets are
superposed on the first structure, using the final multi-feature multiple
alignment. If fit_on_first is False, and position feature was used,
and fit was True, the coordinates will be superposed in the
progressive manner guided by the tree, by the routine that calculates the
inter-molecular distance matrices; this superposition is based only on the
positions of the selected atoms (feature 2), not on other features such as
residue type, secondary, structure, etc. If improve_alignment is
False, it does not make much sense to have fit = True
(use fit_on_first = True).
For local alignments, the matrix offset variable is matrix_offset_3d.
- Feature 3
- is the fractional sidechain accessibility. The pair-wise
residue-residue dissimilarity is calculated by classifying residues into the
buried (
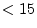 %), semi-exposed, and exposed classes (
%), semi-exposed, and exposed classes (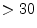 %). The
dissimilarity is 0 for equal classes or if the absolute difference in the
accessibility is less than 5%, 1 for neighboring classes and 2 for
the buried-exposed match. The position-position dissimilarity is the
average residue-residue dissimilarity for comparing all residues from
one group to all residues in the other group (gaps are ignored).
%). The
dissimilarity is 0 for equal classes or if the absolute difference in the
accessibility is less than 5%, 1 for neighboring classes and 2 for
the buried-exposed match. The position-position dissimilarity is the
average residue-residue dissimilarity for comparing all residues from
one group to all residues in the other group (gaps are ignored).
- Feature 4
- is the secondary structure type, distinguishing between helix,
strand, and other. The pair-wise residue-residue dissimilarity is 0 for
equal classes, 1 for `helix' or `strand' matched to `other', and 2 for
`helix' matched to 'strand'. Position-position dissimilarity is calculated
in the same way as for feature 3.
- Feature 5
- is the local conformation. A pair-wise residue-residue score
is DRMSD between the selected atoms (fit_atoms) from the segments of
(2*nsegm + 1) residues centered on the two matched residues. Position-position
dissimilarity is calculated in the same way as for feature 3.
- Feature 6
- is a user specified feature for which a external matrix (in
MODELLER matrix format; see the substitution matrices in the modlib
directory for examples) has to be specified using input_weights_file.
The user can input either a similarity matrix (weights_type = SIMILAR)
or a distance matrix (weights_type = DISTANCE).
- Multiple protein sequence alignment
Aligning multiple sequences is similar to aligning multiple structures,
the difference being that for sequence alignments only the first feature weight
can be non-zero (the other features require coordinates). This is the default
for feature_weights.
- Example: examples/salign/salign_multiple_seq.py
-
# Illustrates the SALIGN multiple sequence alignment
from modeller import *
log.verbose()
env = environ()
env.io.atom_files_directory = ['.', '../atom_files']
aln = alignment(env, file='malign_in.ali')
aln.salign(overhang=30, gap_penalties_1d=(-450, -50),
alignment_type='tree', output='ALIGNMENT')
aln.write(file='malign.ali', alignment_format='PIR')
- Alignment of two sequence blocks
Two blocks of sequences can be aligned using the information contained within
each of the multiple sequence blocks [Martí-Renom et al., 2004].
Pairs of sequence blocks are aligned using SALIGN the same way in which
alignment.align() aligned sequence blocks; to align two blocks, simply
proceed as normal, but set align_block to the number of sequences in
the first block (the rest of the sequences are placed in the second block) and
set alignment_what to BLOCK. Also, since this kind of alignment is
effected only between two blocks, alignment_type is set to PAIRWISE.
- Alignment of protein sequences by their profiles
As for pairwise alignment of sequence blocks, above, two blocks can be
aligned by their profiles. align_block
demarcates the end of the first block and align_what is set to PROFILE
indicating that the blocks will be aligned using their profiles, and
alignment_type is set to PAIRWISE.
The weight matrix for dynamic programming is created by comparing the sequence
information in the two blocks. Two kinds of comparisons can be performed:
- A correlation coefficient of the variation of the the 20 amino acids at
each position (comparison_type is set to PSSM).
- Comparing the residue substitution matrices implied at each position of
the two blocks (comparison_type is set to MAT).
Matrix comparisons are of three types:
taking the maximum, average or correlation coefficient of residue-residue
substitution at the aligned positions (matrix_comparison set to MAX,
AVE or CC).
Profile comparisons are done in similarity space rather than distance space,
so similarity_flag should be set to True. They will also currently
only work with feature 1 (since feature 1 is the only feature which works both
in similarity and in distance space) - the weights for all other features
must be set to zero.
- Example: examples/salign/salign_profile_profile.py
-
# profile-profile alignment using salign
from modeller import *
log.level(1, 0, 1, 1, 1)
env = environ()
aln = alignment(env, file='mega_prune.faa', alignment_format='FASTA')
aln.salign(rr_file='${LIB}/blosum62.sim.mat',
gap_penalties_1d=(-500, 0), output='',
align_block=15, # no. of seqs. in first MSA
align_what='PROFILE',
alignment_type='PAIRWISE',
comparison_type='PSSM', # or 'MAT' (Caution: Method NOT benchmarked
# for 'MAT')
similarity_flag=True, # The score matrix is not rescaled
substitution=True, # The BLOSUM62 substitution values are
# multiplied to the corr. coef.
#output_weights_file='test.mtx', # optional, to write weight matrix
smooth_prof_weight=10.0) # For mixing data with priors
#write out aligned profiles (MSA)
aln.write(file='salign.ali', alignment_format='PIR')
# Make a pairwise alignment of two sequences
aln = alignment(env, file='salign.ali', alignment_format='PIR',
align_codes=('12asA', '1b8aA'))
aln.write(file='salign_pair.ali', alignment_format='PIR')
aln.write(file='salign_pair.pap', alignment_format='PAP')
As stated earlier, all alignment.align() and alignment.align2d() related
commands apply to alignment.salign() too. The example below is a
alignment.salign() equivalent of alignment.align2d() (and
alignment.align()). For a description of the gap_penalties_2d see the
section on alignment.align2d().
- Example: examples/salign/salign_align2d.py
-
# align2d/align using salign
# parameters to be input by the user
# 1. gap_penalties_1d
# 2. gap_penalties_2d
# 3. input alignment file
from modeller import *
log.verbose()
env = environ()
env.io.atom_files_directory = ['../atom_files']
aln = alignment(env, file='align2d_in.ali', align_codes='all')
aln.salign(rr_file='$(LIB)/as1.sim.mat', # Substitution matrix used
output='',
max_gap_length=20,
gap_function=True, # If False then align2d not done
feature_weights=(1., 0., 0., 0., 0., 0.),
gap_penalties_1d=(-100, 0),
gap_penalties_2d=(3.5, 3.5, 3.5, 0.2, 4.0, 6.5, 2.0, 0.0, 0.0),
# d.p. score matrix
#output_weights_file='salign.mtx'
similarity_flag=True) # Ensuring that the dynamic programming
# matrix is not scaled to a difference matrix
aln.write(file='align2d.ali', alignment_format='PIR')
aln.write(file='align2d.pap', alignment_format='PAP')
Caution: The values of gap_penalties_2d have been optimized for
similarity matrices. If using a distance matrix, you will need to derive new
optimized values.
Structure alignments can make use of all the 5 structure/sequence features
as well as the 6th user provided feature matrix. Pairwise alignments of
structures can make use of the constant gap penalties or the environment
dependent gap penalties. Multiple structure alignments are constructed from
pairwise structure alignments.
This section describes the use of SALIGN to produce a single alignment of
multiple structures. If the best output alignment is desired, it is recommended
to run SALIGN in an iterative fashion, to determine the best parameter values.
A utility script is provided for this purpose - see
iterative_structural_align().
The alignment of proteins within a sub-alignment does not change when the
sub-alignment is aligned with another protein or sub-alignment. The pairwise
alignment of sub-alignments is guided by the dendrogram. First, the most
similar pair of proteins are aligned. Second, the next most similar pair of
proteins are aligned, or the third protein is aligned with the sub-alignment
of the first two, as indicated by the dendrogram. This greedy, progressive
procedure requires 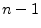 steps to align all
steps to align all 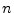 proteins, and each step
requires a pairwise alignment of two sub-alignments.
proteins, and each step
requires a pairwise alignment of two sub-alignments.
If in a multiple alignment, overhangs are to be penalized differently for
the pairs of alignments that create the multiple, auto_overhang can be
set to True. This will ensure that the value of overhang changes as
overhang_factor times the numerical difference in the residues of the
pair. Further, this is only effected if the difference is greater than
overhang_auto_limit.
The dendrogram can be written out in a separate file by specifying the
file name to dendrogram_file.
- Example: examples/salign/salign_multiple_struc.py
-
# Illustrates the SALIGN multiple structure/sequence alignment
from modeller import *
log.verbose()
env = environ()
env.io.atom_files_directory = ['.', '../atom_files']
aln = alignment(env)
for (code, chain) in (('1is4', 'A'), ('1uld', 'D'), ('1ulf', 'B'),
('1ulg', 'B'), ('1is5', 'A')):
mdl = model(env, file=code, model_segment=('FIRST:'+chain, 'LAST:'+chain))
aln.append_model(mdl, atom_files=code, align_codes=code+chain)
for (weights, write_fit, whole) in (((1., 0., 0., 0., 1., 0.), False, True),
((1., 0.5, 1., 1., 1., 0.), False, True),
((1., 1., 1., 1., 1., 0.), True, False)):
aln.salign(rms_cutoff=3.5, normalize_pp_scores=False,
rr_file='$(LIB)/as1.sim.mat', overhang=30,
gap_penalties_1d=(-450, -50),
gap_penalties_3d=(0, 3), gap_gap_score=0, gap_residue_score=0,
dendrogram_file='1is3A.tree',
alignment_type='tree', # If 'progresive', the tree is not
# computed and all structues will be
# aligned sequentially to the first
#ext_tree_file='1is3A_exmat.mtx', # Tree building can be avoided
# if the tree is input
feature_weights=weights, # For a multiple sequence alignment only
# the first feature needs to be non-zero
improve_alignment=True, fit=True, write_fit=write_fit,
write_whole_pdb=whole, output='ALIGNMENT QUALITY')
aln.write(file='1is3A.pap', alignment_format='PAP')
aln.write(file='1is3A.ali', alignment_format='PIR')
# The number of equivalent positions at different RMS_CUTOFF values can be
# computed by changing the RMS value and keeping all feature weights = 0
aln.salign(rms_cutoff=1.0,
normalize_pp_scores=False, rr_file='$(LIB)/as1.sim.mat', overhang=30,
gap_penalties_1d=(-450, -50), gap_penalties_3d=(0, 3),
gap_gap_score=0, gap_residue_score=0, dendrogram_file='1is3A.tree',
alignment_type='progressive', feature_weights=[0]*6,
improve_alignment=False, fit=False, write_fit=True,
write_whole_pdb=False, output='QUALITY')
The weight matrix can be offset at random, many times
over, to generate several `sub-optimal' alignments. The number of sub-optimal
alignments to be output can be specified with n_subopt. Though the matrix
positions at which these offsets are applied cannot be controlled, the user
can choose by how much the matrix will be offset (subopt_offset). The
alignments are written into the file 'suboptimal_alignments.out'
(or 'suboptimal_alignments2d.out' if gap_function is True)
in a simple format. For each such alignment, an 'ALIGNMENT:' line is written,
containing in order
- the number of the alignment (the first alignment is numbered 0, and is
the true or 'optimal' alignment, for reference)
- the start, end and length of the alignment
- the number of aligned positions
- the score of the alignment
- three final fields for internal use
After this the two sequences are written as 'SEQ1' and 'SEQ2', as a simple
mapping from alignment positions to sequences.
Note that subopt_offset should be positive for alignments using distance
matrices (similarity_flag = False) and negative when using similarity
matrices (similarity_flag = True).
Note that because the suboptimal alignments are generated sequentially,
the alignment in memory at the end of the command will be the last (or worst)
suboptimal alignment, not the optimal alignment.
The suboptimal alignment file can be converted into a set of real alignments
using the alignment.get_suboptimals() method.
- Example: examples/salign/salign_subopt.py
-
from modeller import *
log.verbose()
env = environ()
aln = alignment(env, file='fm07254_test.ali', alignment_format='PIR')
aln.salign(feature_weights=(1., 0, 0, 0, 0, 0), gap_penalties_1d=(-450, -50),
n_subopt = 5, subopt_offset = 15)
# Convert suboptimal alignment output file into actual alignments
f = open('suboptimal_alignments.out')
for (n, aln) in enumerate(aln.get_suboptimals(f)):
aln.write(file='fm07254_out%d.ali' % n)
Fix positions:
The user can choose to have certain alignment positions "fixed" by offsetting
the appropriate matrix entries.
This is done by adding a new pseudo sequence to the alignment with the align
code ._fix_pos. The residues of this pseudo sequence are integer values
from 0 through 4 (alternatively, a blank is equivalent to 0).
Any alignment position at which this pseudo sequence contains a '0' is treated
normally; if, however, a non-zero integer is used, the alignment matrix is
offset, generally making the alignment in that position more favorable (and
more so for higher integers). The actual offset values themselves can be
specified by the user by setting the fix_offsets variable.
Note that since SALIGN converts all DP scoring matrices to distance
matrices (unless otherwise specified using similarity_flag), the
values of fix_offsets used in anchoring alignment positions should be
numerically smaller or negative in comparison to the values in the DP matrix.
- Example: examples/salign/salign_fix_positions.py
-
# Demonstrating the use of alignment restraints, only available in
# align2d and salign:
from modeller import *
log.verbose()
env = environ()
# The special alignment entry '_fix_pos' has to be the last entry in the
# alignment array. Its sequence contains characters blank (or 0), 1, 2, 3,
# and 4 at the restrained alignment positions. The residue-residue score from
# the substitution matrix for these positions will be offset by the scalar
# value FIX_OFFSETS[0..4].
aln = alignment(env, file='fix_positions.ali', align_codes=('1leh', '3btoA',
'_fix_pos'))
# fix_offsets specifies the offset corresponding to character ' 1234' in the
# _fix_pos entry in the alignment
# (this offsets unlabeled positions for 0, the ones indicated by 1 by
# 1000, those indicated by 2 by 2000, etc.)
aln.salign(fix_offsets=(0, -10, -20, -30, -40),
gap_penalties_2d=(0, 0, 0, 0, 0, 0, 0, 0, 0), # Any values are
# possible here
local_alignment=False, # Local alignment works, too
gap_penalties_1d=(-600, -400)) # This is best with the default value
# of gap_penalties_2d
# Write it out, the _fix_pos is erased automatically in salign:
aln.write(file='fix_positions_salign.pap', alignment_format='PAP')
External weight matrix: An example of using feature 6.
- Example: examples/salign/salign_external_matrix.py
-
# Reads an external matrix
from modeller import *
log.verbose()
env = environ()
aln = alignment(env, file='1dubA-1nzyA.ali', align_codes='all')
aln.salign(alignment_type='pairwise', output='',
rr_file='$(LIB)/blosum62.sim.mat',
#rr_file='$(LIB)/as1.sim.mat',
#max_gap_length=20,
gap_function=False,
input_weights_file='external.mtx', # External weight matrix
#weights_type='DISTANCE', # type of ext. wgt. mtx
# ensure appropriate gap penalites for the ext. matrix
#feature_weights=(1., 0., 0., 0., 0., 0.), gap_penalties_1d=(30, 26),
#output_weights_file='score.mtx',
feature_weights=(1., 0., 0., 0., 0., 1.),
gap_penalties_1d=(-500, -300))
aln.write(file='output.ali', alignment_format='PIR')
aln.write(file='output.pap', alignment_format='PAP')
Multiple structure alignment according to a user specified dendrogram
The user has the option of inputting an 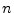 X
X 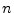 matrix from which a
dendrogram can be inferred. The multiple tree alignment is then confined to
follow this externally input dendrogram. To effect this, specify the name of
the external matrix file with the ext_tree_file variable.
matrix from which a
dendrogram can be inferred. The multiple tree alignment is then confined to
follow this externally input dendrogram. To effect this, specify the name of
the external matrix file with the ext_tree_file variable.
SALIGN makes use of three sets of gap penalties. gap_penalties_1d are for
dynamic programming making use of constant gap penalties. gap_penalties_2d
are when a variable function for gap penalty is used. gap_penalties_3d is
used along with feature 2 only, when structures are aligned by a least squares
fit of their atomic positions.
All SALIGN features produce some measure of residue equivalence (similarity or
distance scores). The scales of these scores differ depending on the feature
used. For optimal usage, gap_penalties_1d should be set appropriately
considering the features used.
Note: If feature 1 is non zero and a similarity substitution matrix is
employed, no matter what other features are also used in conjunction,
gap_penalties_1d should always take on values appropriate to the
substitution matrix used.
For example, if feature 1 is non zero (other features may or may not be
non-zero), and the residue substitution matrix used is the BLOSUM62 similarity
matrix, gap_penalties_1d is set to (-450, -50) and when feature 1 is zero
gap_penalties_1d is set to values appropriate for a distance matrix,
e.g., (2, 3).
A word of caution: gap penalties have not yet been optimized for aligning
sequences by their profiles and for structure alignments.
The gap correction function is
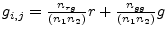 ,
where
,
where 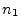 and
and 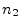 are the number of proteins in the two sub-alignments,
are the number of proteins in the two sub-alignments,
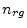 is the number of gap-residue pairs, and
is the number of gap-residue pairs, and 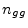 is the number of
gap-gap pairs when comparing protein positions from one sub-alignment with
protein position from the other sub-alignment,
is the number of
gap-gap pairs when comparing protein positions from one sub-alignment with
protein position from the other sub-alignment, 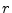 is gap_residue_score
and
is gap_residue_score
and 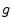 is gap_gap_score. The smaller (even negative) is
gap_gap_score, and the larger is gap_residue_score, the more will
the gaps be aligned with gaps.
is gap_gap_score. The smaller (even negative) is
gap_gap_score, and the larger is gap_residue_score, the more will
the gaps be aligned with gaps.
The alignment.salign() command uses position-position dissimilarity
scores (except when similarity_flag is switched on), as opposed to
similarity scores. This convention applies to all the features, including the
residue-residue similarities read from the rr_file; however, if a residue
type - residue type similarity matrix is read in, it is automatically
converted into the distance matrix by
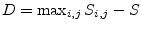 .
In addition, it is also scaled linearly such that the residue-residue
dissimilarity scores range from 0 to 1 (to facilitate weighting this feature
with other features).
.
In addition, it is also scaled linearly such that the residue-residue
dissimilarity scores range from 0 to 1 (to facilitate weighting this feature
with other features).
For each pairwise alignment, the weight matrix 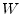 has dimensions
has dimensions 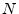 and
and 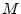 that correspond to the lengths of the sub-alignments to be aligned based on
the weight matrix
that correspond to the lengths of the sub-alignments to be aligned based on
the weight matrix 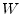 . The dissimilarity score for aligning position
. The dissimilarity score for aligning position 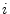 with
position
with
position 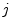 is calculated as
is calculated as
![$ W_{i,j} = \sum_f [ \frac{\omega_f}{\sum_f \omega_f} W^f_{i,j} ] + g_{i,j}$](img142.png) ,
where the sum runs over all selected features
,
where the sum runs over all selected features 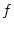 , and
, and 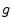 is a function that
may be used to correct the
is a function that
may be used to correct the 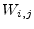 score for the presence of gaps within the
sub-alignments (see below). A feature
score for the presence of gaps within the
sub-alignments (see below). A feature 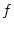 is selected when its weight
is selected when its weight
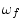 (specified in feature_weights) is non-zero. The matrices
(specified in feature_weights) is non-zero. The matrices
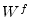 are normalized to have the mean of 0 and standard deviation of 1 when
normalize_pp_scores is True, but it is recommended not to use this
option for now (i.e., use feature_weights to scale the contributions of
the different features to the final
are normalized to have the mean of 0 and standard deviation of 1 when
normalize_pp_scores is True, but it is recommended not to use this
option for now (i.e., use feature_weights to scale the contributions of
the different features to the final 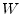 ). The weights of 1 will weigh the
different features approximately evenly (the residue-residue dissimilarities
of feature 1 are scaled to a range from 0 to 1, the position differences of
feature 2 are in angstroms, the fractional solvent accessibility scores of
feature 3 and the secondary structure scores of feature 4 range from 0 to 2,
and the DRMS difference of feature 5 is expressed in angstroms).
). The weights of 1 will weigh the
different features approximately evenly (the residue-residue dissimilarities
of feature 1 are scaled to a range from 0 to 1, the position differences of
feature 2 are in angstroms, the fractional solvent accessibility scores of
feature 3 and the secondary structure scores of feature 4 range from 0 to 2,
and the DRMS difference of feature 5 is expressed in angstroms).
If you enable verbose logging with log.verbose(), there will be more
output in the 'log' file, such as the dendrogram. The dendrogram can also be
written out in a separate file by specifying the file name to
dendrogram_file.
Argument output can contain the following values:
- 'ALIGNMENT': the alignments in the first
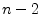 stages of the pairwise
alignment of sub-alignments are written out.
stages of the pairwise
alignment of sub-alignments are written out.
- 'QUALITY': the final alignment is used to obtain pairwise least-squares
superpositions and the corresponding average and minimal numbers of pairs of
aligned residues that are within rms_cutoff Å in all pairs of
aligned structures. These numbers can be used as absolute quality measures
for the final multiple alignment. This option requires the coordinate files
for the aligned proteins.
If write_fit is True, the fitted atom files are written out in their
fitted orientations. For this and other options below, also read the text
above.
If output_weights_file is specified, the dynamic programming weight
matrix is written out into the file. (If it is None, no file is written
out.)
If current_directory is True, the output _pdb.fit files will
be written to the current directory. Otherwise, the output will be in the
directory with the original files.
If write_whole_pdb is True, the whole PDB files are written out;
otherwise only the parts corresponding to the aligned sequences are output.
If fit is False, the initial superposition is not changed. This is
useful when all the structures have to be compared with a given alignment
as is, without changing their relative orientation.
If fit_on_first is True, the structures are fit to the first
structure according to the final alignment before they are written out.
If improve_alignment is False, the initial alignment is not changed,
though the structures may still be superimposed if fit = True. This
is useful when all the structures have to be superimposed with the initial
alignment.




Next: alignment.get_suboptimals() parse
Up: The alignment class: comparison
Previous: alignment.malign3d() align
Contents
Index
Automatic builds
2011-03-29
![]() stages of the progressive multiple alignment, and also
orient all the structures for writing out to atom files with a '_fit.pdb'
extension if write_fit = True. If fit_pdbnam = False, the
PDB filenames in the output alignment file will not have the '_fit.pdb'
extensions. Thus, feature 2 needs to be selected by
feature_weight[2]
stages of the progressive multiple alignment, and also
orient all the structures for writing out to atom files with a '_fit.pdb'
extension if write_fit = True. If fit_pdbnam = False, the
PDB filenames in the output alignment file will not have the '_fit.pdb'
extensions. Thus, feature 2 needs to be selected by
feature_weight[2] ![]() if you wish to write out the structures
superposed according to the tree-following procedure; also, fit_on_first
must be False, otherwise the structures are written out superposed on the
first structure according to the final alignment (see also below).
if you wish to write out the structures
superposed according to the tree-following procedure; also, fit_on_first
must be False, otherwise the structures are written out superposed on the
first structure according to the final alignment (see also below).
![]() does not
generally correspond to the alignment calculated based on
does not
generally correspond to the alignment calculated based on ![]() . Therefore, the
multiply superposed structures are not necessarily superposed based on the
final multiple alignment produced by alignment.salign(). If you wish such
a superposition, you can use alignment.malign3d() with fit = False
and write_fit = True (the meaning of fit is different between
alignment.salign() and alignment.malign3d()).
. Therefore, the
multiply superposed structures are not necessarily superposed based on the
final multiple alignment produced by alignment.salign(). If you wish such
a superposition, you can use alignment.malign3d() with fit = False
and write_fit = True (the meaning of fit is different between
alignment.salign() and alignment.malign3d()).