


Next: About this document ...
Up: MODELLER A Program for
Previous: Bibliography
Contents
- A
- Mathematical forms of restraints
- accessibility_type
- model.write_data() write
| Restraints.make() make
- actions
- conjugate_gradients() optimize
| molecular_dynamics() optimize
| actions.write_structure() write
| actions.charmm_trajectory() write
- actions.charmm_trajectory()
- automodel.get_optimize_actions() get
| model.write_psf() write
| actions.charmm_trajectory() write
- actions.trace()
- automodel.trace_output control
| conjugate_gradients() optimize
| actions.trace() write
| TOP to Python correspondence
- actions.write_structure()
- conjugate_gradients() optimize
| actions.write_structure() write
| actions.charmm_trajectory() write
- add
- Restraints.add() add
| selection.add() add
- add_leading_gaps
- Residue.add_leading_gaps() add
- add_trailing_gaps
- Residue.add_trailing_gaps() add
- align
- alignment.align() align
- align2d
- alignment.align2d() align
- align3d
- alignment.align3d() align
- align3d_repeat
- alignment.align3d() align
- align3d_trf
- alignment.align3d() align
| Features of proteins used
- align_alignment
- alignment.write() write
- align_block
- alignment.write() write
| alignment.align() align
| alignment.malign() align
| Alignment of protein sequences
| alignment.segment_matching() align
- align_code
- Chain.write() write
| Chain.atom_file_and_code() get
- align_codes
- alignment.append() read
| alignment.append_model() copy
- align_what
- alignment.align() align
| Alignment of protein sequences
- alignment
- Alignment file
| alignment() create
- alignment class
- The alignment class: comparison
- alignment files
- format
- Alignment file (PIR)
- reading
- alignment.append() read
- writing
- alignment.write() write
- alignment Structure class
- The Structure class: a
- alignment()
- alignment() create
- alignment.align()
- alignment.write() write
| alignment.align() align
| alignment.align2d() align
| alignment.malign() align
| alignment.consensus() consensus
| alignment.align3d() align
| alignment.salign() align
| Alignment of protein sequences
| Alignment of protein structures
| sequence_db.filter() cluster
| TOP to Python correspondence
- alignment.align2d
- Residue.curvature Mainchain
- alignment.align2d()
- Coordinate files and derivative
| alignment.write() write
| alignment.align() align
| alignment.align2d() align
| alignment.salign() align
| Alignment of protein structures
| TOP to Python correspondence
- alignment.align3d()
- selection.superpose() superpose
| alignment.align() align
| alignment.align3d() align
| alignment.malign3d() align
| alignment.salign() align
| Features of proteins used
| TOP to Python correspondence
- alignment.append()
- Changes since release 9v7
| alignment() create
| alignment.append() read
| alignment.read_one() read
| alignment.compare_with() compare
| alignment.write() write
| alignment.describe() describe
| profile.build() Build
| Flowchart of comparative modeling
| TOP to Python correspondence
- alignment.append_model()
- selection.mutate() mutate
| alignment.append_model() copy
| Flowchart of comparative modeling
| TOP to Python correspondence
- alignment.append_profile()
- alignment.append_profile() add
| TOP to Python correspondence
- alignment.append_sequence()
- model.build_sequence() build
| alignment.append_sequence() add
- alignment.check()
- Alignment file
| Running MODELLER
| Fully automated alignment and
| Frequently asked questions (FAQ)
| alignment.check() check
| Structure.reread() reread
| Flowchart of comparative modeling
| TOP to Python correspondence
- alignment.check_sequence_structure()
- alignment.check_sequence_structure() check
| alignment.check() check
- alignment.check_structure_structure()
- alignment.check_structure_structure() check
| alignment.check() check
- alignment.clear()
- alignment.clear() delete
- alignment.comments
- alignment.comments alignment
- alignment.compare_sequences()
- environ.dendrogram() clustering
| environ.principal_components() clustering
| alignment.id_table() calculate
| alignment.compare_sequences() compare
| TOP to Python correspondence
- alignment.compare_structures()
- Coordinate files and derivative
| environ.dendrogram() clustering
| environ.principal_components() clustering
| alignment.compare_structures() compare
| TOP to Python correspondence
- alignment.compare_with()
- alignment.compare_with() compare
| alignment.align() align
| TOP to Python correspondence
- alignment.consensus()
- alignment.write() write
| alignment.consensus() consensus
| TOP to Python correspondence
- alignment.describe()
- alignment.describe() describe
| TOP to Python correspondence
- alignment.edit()
- alignment.edit() edit
| TOP to Python correspondence
- alignment.get_suboptimals()
- Changes since release 9v8
| Sub-optimal alignments
| alignment.get_suboptimals() parse
- alignment.id_table()
- environ.dendrogram() clustering
| environ.principal_components() clustering
| alignment.id_table() calculate
| alignment.compare_sequences() compare
| TOP to Python correspondence
- alignment.malign()
- alignment.align() align
| alignment.malign() align
| alignment.malign3d() align
| alignment.salign() align
| Features of proteins used
| sequence_db.search() search
| TOP to Python correspondence
- alignment.malign3d()
- Coordinate files and derivative
| model.transfer_xyz() copy
| alignment.align() align
| alignment.compare_structures() compare
| alignment.malign3d() align
| alignment.salign() align
| Features of proteins used
| Flowchart of comparative modeling
| TOP to Python correspondence
- alignment.read_one()
- alignment.read_one() read
| modfile.File() open
- alignment.salign
- Residue.curvature Mainchain
- alignment.salign()
- Changes since release 9v8
| alignment.align() align
| alignment.align2d() align
| alignment.malign() align
| alignment.align3d() align
| alignment.malign3d() align
| alignment.salign() align
| Features of proteins used
| Alignment of protein structures
| Useful SALIGN information and
| alignment.get_suboptimals() parse
| The salign module: high-level
| iterative_structural_align() obtain
| TOP to Python correspondence
- alignment.segment_matching()
- alignment.segment_matching() align
| TOP to Python correspondence
- alignment.to_profile
- environ.make_pssmdb() Create
- alignment.to_profile()
- alignment.to_profile() convert
| profile() create
| profile.to_alignment() profile
| profile.scan() Compare
| profile.build() Build
| Profile file
| TOP to Python correspondence
- alignment.write()
- selection.mutate() mutate
| alignment.write() write
| alignment.align() align
| alignment.align2d() align
| alignment.consensus() consensus
| alignment.malign3d() align
| Chain.write() write
| profile.build() Build
| sequence_db.search() search
| TOP to Python correspondence
- alignment_features
- alignment.write() write
- alignment_format
- alignment.write() write
- alignment_type
- Alignment of protein sequences
| Alignment of protein structures
- alignment_what
- Alignment of protein sequences
- allhmodel
- allhmodel() prepare
- allhmodel class
- allhmodel reference
- allhmodel()
- allhmodel() prepare
- allow_alternates
- Changes since release 9v7
| alignment.append() read
- aln
- automodel.special_restraints() add
| model.get_insertions() return
| model.get_deletions() return
| model.patch_ss_templates() guess
| model.transfer_xyz() copy
| model.res_num_from() residue
| model.color() color
| Restraints.make() make
| selection.superpose() superpose
| alignment.compare_with() compare
| sequence_db.search() search
- aln_base_filename
- profile.scan() Compare
- alnfile
- automodel() prepare
| loopmodel() prepare
- alnseq
- model.generate_topology() generate
- alpha
- alpha() make
- alpha()
- alpha() make
- angular_step_size
- density.grid_search() dock
- append
- Topology.append() append
| Parameters.append() append
| Restraints.append() read
| group_restraints.append() read
| alignment.append() read
- append_model
- alignment.append_model() copy
- append_profile
- alignment.append_profile() add
- append_sequence
- alignment.append_sequence() add
- asgl_output
- selection.energy() evaluate
| alignment.compare_structures() compare
- assess_dope
- selection.assess_dope() assess
- assess_dopehr
- selection.assess_dopehr() assess
- assess_ga341
- model.assess_ga341() assess
- assess_methods
- Getting a very fast
| automodel() prepare
- assess_normalized_dope
- model.assess_normalized_dope() assess
- atmsel
- Restraints.make() make
| Restraints.pick() pick
- atmsel1
- Restraints.make_distance() make
- atmsel2
- Restraints.make_distance() make
- Atom class
- The Atom class: a
- atom classes
- Loop modeling method
- Atom.accessibility
- model.write_data() write
| Atom.accessibility atomic
- Atom.biso
- model.write_data() write
| model.make_region() define
| Atom.biso isotropic
- Atom.charge
- Atom.charge electrostatic
- Atom.dvx
- Atom.dvx objective
- Atom.mass
- Atom.mass mass
- Atom.name
- Atom.name PDB
- Atom.occ
- Atom.occ occupancy
- Atom.residue
- Atom.residue residue
- Atom.type
- Atom.type CHARMM
- Atom.vx
- Atom.vx x
- atom1
- sheet() make
- atom2
- sheet() make
- atom_accessibility
- model.make_region() define
- atom_file
- Chain.write() write
| Chain.atom_file_and_code() get
- atom_file_and_code
- Chain.atom_file_and_code() get
- atom_files
- alignment.append() read
| alignment.append_model() copy
- atom_ids
- Specification of pseudo atoms
| Restraints.unpick() unselect
- atom_range
- model.atom_range() return
- AtomType class
- The AtomType class: a
- AtomType.element
- AtomType.element element
- AtomType.mass
- AtomType.mass atomic
- AtomType.name
- AtomType.name CHARMM
- auto_align
- automodel.auto_align() generate
- auto_overhang
- Alignment of protein structures
- automodel
- automodel() prepare
- automodel class
- automodel reference
- automodel()
- automodel() prepare
| allhmodel() prepare
| loopmodel() prepare
- automodel.assess_methods
- loopmodel() prepare
- automodel.auto_align()
- Fully automated alignment and
| automodel.auto_align() generate
- automodel.cluster()
- automodel.cluster() cluster
- automodel.deviation
- automodel.rand_method control
| Flowchart of comparative modeling
- automodel.ending_model
- Simple usage
| automodel.starting_model first
| automodel.ending_model last
| loopmodel.loop.ending_model last
| selection.assess_dope() assess
- automodel.final_malign3d
- automodel.final_malign3d final
| loopmodel.loop.write_selection_only write
| Flowchart of comparative modeling
- automodel.generate_method
- Using your own initial
| automodel.generate_method control
- automodel.get_model_filename()
- automodel.get_model_filename() get
| loopmodel.get_loop_model_filename() get
- automodel.get_optimize_actions()
- automodel.trace_output control
| automodel.get_optimize_actions() get
| automodel.get_refine_actions() get
- automodel.get_refine_actions()
- automodel.trace_output control
| automodel.get_refine_actions() get
- automodel.initial_malign3d
- Building a model from
| automodel.initial_malign3d initial
| model.transfer_xyz() copy
| alignment.check_structure_structure() check
- automodel.library_schedule
- Changing the default optimization
| automodel() prepare
| automodel.library_schedule select
| automodel.very_fast() request
| loopmodel.loop.library_schedule select
- automodel.make()
- Simple usage
| Getting a very fast
| Accessing output data after
| Fully automated alignment and
| automodel.outputs all
| automodel.make() build
- automodel.max_ca_ca_distance
- automodel.max_ca_ca_distance Distance
| automodel.max_n_o_distance Distance
| automodel.max_sc_mc_distance Distance
| automodel.max_sc_sc_distance Distance
| automodel.very_fast() request
- automodel.max_molpdf
- Changing the default optimization
| automodel.max_molpdf objective
- automodel.max_n_o_distance
- automodel.max_ca_ca_distance Distance
| automodel.max_n_o_distance Distance
- automodel.max_sc_mc_distance
- automodel.max_ca_ca_distance Distance
| automodel.max_sc_mc_distance Distance
- automodel.max_sc_sc_distance
- automodel.max_ca_ca_distance Distance
| automodel.max_sc_sc_distance Distance
- automodel.max_var_iterations
- Changing the default optimization
| automodel.max_var_iterations select
| loopmodel.loop.max_var_iterations select
- automodel.md_level
- Changing the default optimization
| automodel.md_level control
| automodel.very_fast() request
| loopmodel.loop.md_level control
- automodel.nonstd_restraints()
- Including water molecules, HETATM
| Changes since release 9v7
| automodel.nonstd_restraints() add
| Block (BLK) residues with
| Flowchart of comparative modeling
- automodel.outputs
- Accessing output data after
| automodel.outputs all
| loopmodel.loop.outputs all
- automodel.rand_method
- automodel() prepare
| automodel.rand_method control
| automodel.very_fast() request
- automodel.repeat_optimization
- Changing the default optimization
| automodel.repeat_optimization number
- automodel.select_atoms()
- Refining only part of
| automodel.select_atoms() select
- automodel.special_patches()
- Including disulfide bridges
| Building multi-chain models with
| automodel.special_patches() add
| Flowchart of comparative modeling
- automodel.special_restraints()
- Adding additional restraints to
| Building multi-chain models with
| automodel.special_restraints() add
| Flowchart of comparative modeling
- automodel.starting_model
- Simple usage
| automodel.starting_model first
| automodel.ending_model last
| loopmodel.loop.starting_model first
- automodel.trace_output
- automodel.trace_output control
| automodel.get_optimize_actions() get
- automodel.use_parallel_job()
- automodel.use_parallel_job() parallelize
- automodel.user_after_single_model()
- Building multi-chain models with
| automodel.user_after_single_model() analyze
| loopmodel.user_after_single_loop_model() analyze
- automodel.very_fast()
- Getting a very fast
| automodel.max_ca_ca_distance Distance
| automodel.very_fast() request
- automodel.write_intermediates
- automodel.write_intermediates write
- B
- Mathematical forms of restraints
- base_align_codes
- alignment.edit() edit
- basis_pdf_weight
- Restraints.make() make
- basis_relative_weight
- Restraints.make() make
- best_docked_models
- density.grid_search() dock
- bibliography
- MODELLER bibliography
- BLOCK residues
- Block (BLK) residues with
- breakpoint
- Controlling breakpoints and the
- bug reports
- Bug reports
- build
- model.build() build
| profile.build() Build
- build_ini_loop
- loopmodel.build_ini_loop() create
- build_method
- model.build() build
- build_sequence
- model.build_sequence() build
- by_residue
- selection.by_residue() make
- cap_atom_shift
- molecular_dynamics() optimize
| Molecular dynamics
- ccf_func_type
- density.read() read
- ccmatrix_offset
- profile.scan() Compare
- Chain class
- The Chain class: a
- Chain.atom_file_and_code()
- Chain.write() write
| Chain.atom_file_and_code() get
- Chain.atoms
- Chain.atoms all
- Chain.filter()
- model.make_chains() Fetch
| Chain.filter() check
| Chain.write() write
| Chain.atom_file_and_code() get
- Chain.join()
- Changes since release 9v7
| Chain.join() join
- Chain.name
- Chain.name chain
- Chain.residues
- Chain.residues all
- Chain.write()
- model.make_chains() Fetch
| Chain.filter() check
| Chain.write() write
| Chain.atom_file_and_code() get
- chains_list
- sequence_db.read() read
| sequence_db.write() write
- change
- selection.rotate_dihedrals() change
- charmm_trajectory
- actions.charmm_trajectory() write
- check
- alignment.check() check
- check_profile
- profile.build() Build
- check_sequence_structure
- alignment.check_sequence_structure() check
- check_structure_structure
- alignment.check_structure_structure() check
- chop_nonstd_termini
- Chain.filter() check
| Chain.write() write
- cispeptide
- cispeptide() creates
- cispeptide()
- Restraints.add() add
| Restraints.unpick() unselect
| cispeptide() creates
- classes
- group_restraints() create
| group_restraints.append() read
- clean_sequences
- sequence_db.read() read
- clear
- Topology.clear() clear
| Parameters.clear() clear
| Restraints.clear() delete
| alignment.clear() delete
- clear_topology
- model.clear_topology() clear
- cluster
- automodel.cluster() cluster
- cluster_cut
- automodel.cluster() cluster
| model.transfer_xyz() copy
- cluster_method
- model.transfer_xyz() copy
- color
- model.color() color
- comment
- Chain.write() write
- Communicator.get_data()
- Parallel job support
| job() create
| Communicator.send_data() send
| Communicator.get_data() get
- Communicator.send_data()
- Parallel job support
| job() create
| job.queue_task() submit
| Communicator.send_data() send
| Communicator.get_data() get
| slave.run_cmd() run
- compactness
- model.assess_ga341() assess
- compare_mode
- alignment.compare_structures() compare
- compare_sequences
- alignment.compare_sequences() compare
- compare_structures
- alignment.compare_structures() compare
- compare_with
- alignment.compare_with() compare
- comparison_type
- Alignment of protein sequences
- complete_pdb
- model.write_psf() write
| complete_pdb() read
- complete_pdb()
- Changes since release 9v8
| model.read() read
| model.build_sequence() build
| model.write() write
| selection.assess_dope() assess
| Chain.join() join
| complete_pdb() read
- compressed files
- Reading or writing files
- condense
- Restraints.condense() remove
- conjugate_gradients
- conjugate_gradients() optimize
- conjugate_gradients()
- automodel.max_var_iterations select
| Rigid bodies
| Restraints.make_distance() make
| conjugate_gradients() optimize
| quasi_newton() optimize
| molecular_dynamics() optimize
| actions.write_structure() write
| actions.trace() write
| actions.charmm_trajectory() write
| complete_pdb() read
| Flowchart of comparative modeling
| TOP to Python correspondence
- consensus
- alignment.consensus() consensus
- contact_shell
- Feature types
- convert
- sequence_db.convert() convert
- correls
- Mathematical forms of restraints
- Coulomb
- Block (BLK) residues with
| Static and dynamic restraints
| Mathematical forms of restraints
| Rigid bodies
| The energy_data class: objective
| energy_data.contact_shell nonbond
| energy_data.dynamic_coulomb calculate
| energy_data.coulomb_switch Coulomb
| energy_data.relative_dielectric relative
| model.read() read
| Restraints.make() make
| physical.values() create
| Function
| Coulomb restraint
- csrfile
- automodel() prepare
- current_directory
- alignment.malign3d() align
| Useful SALIGN information and
- cutoff
- gbsa.Scorer() create
- data_file
- sequence_db.search() search
- debug_function
- selection.debug_function() test
- debug_function_cutoff
- selection.debug_function() test
- default
- modfile.default() generate
- delete
- modfile.delete() delete
- delta
- Mathematical forms of restraints
- dendrogram
- environ.dendrogram() clustering
- dendrogram_file
- Alignment of protein structures
| Useful SALIGN information and
- density
- density() create
- density class
- The density class: handling
- density()
- density() create
- density.grid_search()
- The density class: handling
| density() create
| density.read() read
| density.grid_search() dock
| TOP to Python correspondence
- density.read()
- Changes since release 9v7
| density() create
| density.resolution Map
| density.voxel_size Map
| density.read() read
| density.grid_search() dock
- density.resolution
- density.resolution Map
- density.voxel_size
- density.voxel_size Map
- density_type
- density.read() read
| density.grid_search() dock
- describe
- alignment.describe() describe
- detailed_debugging
- selection.debug_function() test
- deviation
- automodel() prepare
| Restraints.symmetry.report() report
| selection.randomize_xyz() randomize
- dih_lib_only
- Restraints.make() make
- Dihedral class
- The Dihedral class: a
- Dihedral.atoms
- Dihedral.atoms atoms
- Dihedral.dihclass
- Dihedral.dihclass integer
- Dihedral.value
- Dihedral.value current
- dihedrals
- selection.rotate_dihedrals() change
- distance_rsr_model
- Restraints.make_distance() make
- disulfide bond restraints
- Including disulfide bridges
- dnr_accpt_lib
- model.write_data() write
- dope_loopmodel
- dope_loopmodel() prepare
- dope_loopmodel class
- dope_loopmodel reference
- dope_loopmodel()
- dope_loopmodel() prepare
| The gbsa module: implicit
- dopehr_loopmodel class
- dopehr_loopmodel reference
- dr
- saxsdata.ini_saxs() Initialization
- dr_exp
- saxsdata.ini_saxs() Initialization
- e_native_comb
- model.assess_ga341() assess
- e_native_pair
- model.assess_ga341() assess
- e_native_surf
- model.assess_ga341() assess
- edat
- Key for command descriptions
| environ.edat default
| conjugate_gradients() optimize
| molecular_dynamics() optimize
- edit
- alignment.edit() edit
- edit_align_codes
- alignment.edit() edit
- edit_file_ext
- alignment.malign3d() align
- electrostatics
- see Coulomb
- em_fit_output_file
- density.grid_search() dock
- em_map_size
- density.read() read
- em_pdb_name
- density.grid_search() dock
- end
- model.atom_range() return
| model.residue_range() return
- end_of_file
- alignment.append() read
- energy
- selection.energy() evaluate
- energy_data
- Key for command descriptions
| energy_data() create
- energy_data class
- The energy_data class: objective
- energy_data()
- energy_data() create
- energy_data.contact_shell
- energy_data.contact_shell nonbond
| energy_data.update_dynamic nonbond
| energy_data.nlogn_use select
| Restraints.make() make
| selection.assess_dope() assess
| conjugate_gradients() optimize
| gbsa.Scorer() create
| Atomic density
| TOP to Python correspondence
- energy_data.coulomb_switch
- energy_data.contact_shell nonbond
| energy_data.coulomb_switch Coulomb
| gbsa.Scorer() create
| TOP to Python correspondence
- energy_data.covalent_cys
- energy_data.covalent_cys use
| TOP to Python correspondence
- energy_data.density
- Static and dynamic restraints
- energy_data.dynamic_coulomb
- Static and dynamic restraints
| energy_data.contact_shell nonbond
| energy_data.dynamic_coulomb calculate
| Restraints.make() make
| TOP to Python correspondence
- energy_data.dynamic_lennard
- Static and dynamic restraints
| energy_data.contact_shell nonbond
| energy_data.dynamic_lennard calculate
| Restraints.make() make
| TOP to Python correspondence
- energy_data.dynamic_modeller
- Block (BLK) residues with
| Static and dynamic restraints
| energy_data.dynamic_modeller calculate
| energy_data.covalent_cys use
| model.group_restraints all
| The group_restraints class: restraints
| TOP to Python correspondence
- energy_data.dynamic_sphere
- Static and dynamic restraints
| energy_data.contact_shell nonbond
| energy_data.sphere_stdv soft-sphere
| energy_data.dynamic_sphere calculate
| Restraints.make() make
| TOP to Python correspondence
- energy_data.energy_terms
- energy_data.energy_terms user-defined
| gbsa.Scorer() create
| User-defined energy terms
- energy_data.excl_local
- energy_data.excl_local exclude
| Restraints.make_distance() make
| TOP to Python correspondence
- energy_data.lennard_jones_switch
- energy_data.contact_shell nonbond
| energy_data.lennard_jones_switch Lennard-Jones
| TOP to Python correspondence
- energy_data.nlogn_use
- energy_data.nlogn_use select
| conjugate_gradients() optimize
| TOP to Python correspondence
- energy_data.nonbonded_sel_atoms
- Refining only part of
| energy_data.nonbonded_sel_atoms control
| Restraints.make() make
| Restraints.pick() pick
| selection.assess_dope() assess
| TOP to Python correspondence
- energy_data.radii_factor
- energy_data.contact_shell nonbond
| energy_data.radii_factor scale
| model.write_data() write
| TOP to Python correspondence
- energy_data.relative_dielectric
- energy_data.relative_dielectric relative
| Coulomb restraint
| TOP to Python correspondence
- energy_data.saxsdata
- Static and dynamic restraints
- energy_data.sphere_stdv
- energy_data.sphere_stdv soft-sphere
| energy_data.dynamic_sphere calculate
| TOP to Python correspondence
- energy_data.update_dynamic
- energy_data.contact_shell nonbond
| energy_data.update_dynamic nonbond
| conjugate_gradients() optimize
| molecular_dynamics() optimize
| TOP to Python correspondence
- env.io.hetatm
- Including water molecules, HETATM
| model.read() read
- env.io.hydrogen
- Including water molecules, HETATM
| Building an all hydrogen
- env.io.water
- Including water molecules, HETATM
- environ
- environ() create
- environ class
- The environ class: MODELLER
- environ()
- Simple usage
| Frequently asked questions (FAQ)
| environ() create
| TOP to Python correspondence
- environ.dendrogram()
- Changes since release 9v8
| environ.dendrogram() clustering
| model.transfer_xyz() copy
| alignment.id_table() calculate
| alignment.compare_structures() compare
| sequence_db.search() search
| TOP to Python correspondence
- environ.edat
- Key for command descriptions
| environ.edat default
| energy_data() create
- environ.io
- Key for command descriptions
| environ.io default
| io_data() create
- environ.libs
- environ.libs MODELLER
- environ.make_pssmdb()
- environ.make_pssmdb() Create
| profile.scan() Compare
- environ.principal_components()
- Changes since release 9v7
| environ.principal_components() clustering
| alignment.id_table() calculate
| alignment.compare_structures() compare
| TOP to Python correspondence
- environ.schedule_scale
- Changing the default optimization
| environ.schedule_scale energy
| schedule() create
- environ.system()
- environ.system() execute
| TOP to Python correspondence
- equilibrate
- molecular_dynamics() optimize
| Molecular dynamics
- eqvdst
- alignment.check_structure_structure() check
- errors
- EOFError
- Controlling breakpoints and the
- FileFormatError
- Controlling breakpoints and the
| model.read() read
| alignment.append() read
| alignment.read_one() read
- handling and recovering
- Controlling breakpoints and the
- IndexError
- Controlling breakpoints and the
- IOError
- Controlling breakpoints and the
- MemoryError
- Controlling breakpoints and the
- ModellerError
- Controlling breakpoints and the
| model.assess_ga341() assess
- NetworkError
- Communicator.send_data() send
- NotImplementedError
- Controlling breakpoints and the
| Restraints.spline() approximate
- RemoteError
- Communicator.send_data() send
- SequenceMismatchError
- alignment.append() read
| alignment.read_one() read
- standard Python
- Controlling breakpoints and the
- StatisticsError
- profile.build() Build
- TypeError
- Controlling breakpoints and the
- ValueError
- Controlling breakpoints and the
| Restraints.spline() approximate
| Chain.join() join
- ZeroDivisionError
- Controlling breakpoints and the
- exclude_distance
- Restraints.make_distance() make
- excluded pairs
- Excluded pairs
- excluded_pair
- Excluded pairs
- exit_stage
- automodel.make() build
- ext_tree_file
- Alignments using external restraints
- extend_by_residue
- selection.extend_by_residue() extend
- extension
- model.get_insertions() return
| model.get_deletions() return
| selection.extend_by_residue() extend
- factor
- Mathematical forms of restraints
- fast_search
- sequence_db.search() search
- fast_search_cutoff
- sequence_db.search() search
- feature
- Mathematical forms of restraints
| Restraints.spline() approximate
- feature_weight
- Features of proteins used
- feature_weights
- Alignment of protein sequences
| Useful SALIGN information and
- features.angle
- Feature types
- features.density
- Feature types
- features.dihedral
- Feature types
- features.dihedral_diff
- Feature types
- features.distance
- Feature types
- features.minimal_distance
- Feature types
- features.solvent_access
- Feature types
- features.x_coordinate
- Feature types
- features.y_coordinate
- Feature types
- features.z_coordinate
- Feature types
- fh
- schedule.write() write
- File
- environ.principal_components() clustering
| Topology.append() append
| Parameters.append() append
| model.write() write
| model.write_data() write
| group_restraints.append() read
| alignment.read_one() read
| alignment.write() write
| alignment.segment_matching() align
| Structure.write() write
| Chain.write() write
| density.read() read
| modfile.File() open
- file naming
- File naming
- file types
- File types
- file_exists
- modfile.inquire() check
- file_ext
- alignment.segment_matching() align
| modfile.default() generate
- file_id
- alignment.segment_matching() align
| modfile.default() generate
- filename
- saxsdata.ini_saxs() Initialization
| complete_pdb() read
- filepattern
- actions.write_structure() write
- filter
- Chain.filter() check
| sequence_db.filter() cluster
- filter_type
- density.read() read
- filter_values
- density.read() read
- first
- actions.write_structure() write
| actions.charmm_trajectory() write
- fit
- selection.superpose() superpose
| alignment.write() write
| alignment.align2d() align
| alignment.compare_structures() compare
| alignment.align3d() align
| alignment.malign3d() align
| Features of proteins used
| Useful SALIGN information and
- fit_atoms
- alignment.compare_structures() compare
| alignment.align3d() align
| alignment.malign3d() align
| Features of proteins used
- fit_on_first
- Features of proteins used
| Useful SALIGN information and
- fit_pdbnam
- Features of proteins used
- fix_offsets
- Alignments using external restraints
- form
- Restraints.spline() approximate
- format
- Chain.write() write
- forms.cosine
- Mathematical forms of restraints
- forms.coulomb
- Mathematical forms of restraints
- forms.factor
- Mathematical forms of restraints
- forms.gaussian
- Mathematical forms of restraints
- forms.lennard_jones
- Mathematical forms of restraints
- forms.lower_bound
- Mathematical forms of restraints
- forms.multi_binormal
- Mathematical forms of restraints
- forms.multi_gaussian
- Mathematical forms of restraints
- forms.nd_spline
- Mathematical forms of restraints
- forms.spline
- Mathematical forms of restraints
- forms.upper_bound
- Mathematical forms of restraints
- frequently asked questions
- Frequently asked questions (FAQ)
- gap_function
- alignment.salign() align
| Alignment of protein structures
| Sub-optimal alignments
| Residue.curvature Mainchain
- gap_gap_score
- Gap penalties and correcting
- gap_penalties_1d
- alignment.align() align
| alignment.align2d() align
| alignment.malign() align
| alignment.salign() align
| Gap penalties and correcting
| profile.scan() Compare
| sequence_db.search() search
| sequence_db.filter() cluster
| Profile file
- gap_penalties_2d
- alignment.align2d() align
| Alignment of protein structures
| Gap penalties and correcting
- gap_penalties_3d
- alignment.align3d() align
| alignment.malign3d() align
| Features of proteins used
| Gap penalties and correcting
- gap_residue_score
- Gap penalties and correcting
- gapdist
- alignment.check_sequence_structure() check
- gaps_in_target
- profile.build() Build
- GB/SA scoring
- gbsa.Scorer() create
- gbsa.Scorer()
- Block (BLK) residues with
| gbsa.Scorer() create
- generate_topology
- model.generate_topology() generate
- get_aligned_residue
- Residue.get_aligned_residue() get
- get_data
- Communicator.get_data() get
- get_deletions
- model.get_deletions() return
- get_insertions
- model.get_insertions() return
- get_leading_gaps
- Residue.get_leading_gaps() get
- get_loop_model_filename
- loopmodel.get_loop_model_filename() get
- get_model_filename
- automodel.get_model_filename() get
- get_num_equiv
- Sequence.get_num_equiv() get
- get_optimize_actions
- automodel.get_optimize_actions() get
- get_range
- Restraints.spline() approximate
- get_refine_actions
- automodel.get_refine_actions() get
- get_sequence_identity
- Sequence.get_sequence_identity() get
- get_suboptimals
- alignment.get_suboptimals() parse
- get_trailing_gaps
- Residue.get_trailing_gaps() get
- grid_search
- density.grid_search() dock
- group
- Restraints.spline() approximate
- group_restraints
- group_restraints() create
- group_restraints class
- The group_restraints class: restraints
- group_restraints()
- Block (BLK) residues with
| model.group_restraints all
| group_restraints() create
| TOP to Python correspondence
- group_restraints.append()
- group_restraints() create
| group_restraints.append() read
| TOP to Python correspondence
- guide_factor
- molecular_dynamics() optimize
- guide_time
- molecular_dynamics() optimize
- high
- Mathematical forms of restraints
- highderiv
- Mathematical forms of restraints
- host
- job() create
- hot_atoms
- selection.hot_atoms() atoms
- id1
- modfile.default() generate
- id2
- modfile.default() generate
- id_table
- alignment.id_table() calculate
- improve_alignment
- Features of proteins used
| Useful SALIGN information and
- info.bindir
- info.bindir MODELLER
- info.build_date
- info.version the
| info.build_date the
- info.debug
- info.debug this
- info.exe_type
- info.exe_type the
- info.jobname
- info.jobname name
| job() create
- info.time_mark()
- info.time_mark() print
| TOP to Python correspondence
- info.version
- info.version the
- info.version_info
- info.version the
| info.version_info the
- ini_saxs
- saxsdata.ini_saxs() Initialization
- inifile
- automodel() prepare
- inimodel
- loopmodel() prepare
- init_velocities
- molecular_dynamics() optimize
- initialize_xyz
- model.build() build
- input_profile_file
- alignment.align2d() align
- input_weights_file
- Features of proteins used
- inquire
- modfile.inquire() check
- installation
- Obtaining and installing the
- intersegment
- Restraints.make() make
- io
- Key for command descriptions
| environ.io default
| model.generate_topology() generate
| alignment.append() read
| alignment.edit() edit
- io_data
- io_data() create
- io_data class
- The io_data class: coordinate
- io_data()
- Key for command descriptions
| io_data() create
| model.read() read
- io_data.atom_files_directory
- Simple usage
| Coordinate files and derivative
| io_data.atom_files_directory search
| Alignment file (PIR)
| TOP to Python correspondence
- io_data.hetatm
- Block (BLK) residues with
| io_data() create
| io_data.hetatm whether
| TOP to Python correspondence
- io_data.hydrogen
- io_data() create
| io_data.hydrogen whether
| TOP to Python correspondence
- io_data.water
- io_data() create
| io_data.hetatm whether
| io_data.water whether
| TOP to Python correspondence
- iterative_structural_align
- iterative_structural_align() obtain
- iterative_structural_align()
- Alignment of protein structures
| iterative_structural_align() obtain
- job
- job() create
- job class
- Parallel job support
- job()
- job() create
| sge_pe_job() create
| sge_qsub_job() create
- job.queue_task()
- Parallel job support
| job() create
| job.queue_task() submit
- job.run_all_tasks()
- Parallel job support
| job() create
| job.queue_task() submit
| job.run_all_tasks() run
| job.yield_tasks_unordered() run
- job.slave_startup_commands
- job() create
| job.slave_startup_commands Slave
- job.start()
- Parallel job support
| job() create
| job.start() start
- job.yield_tasks_unordered()
- Parallel job support
| job() create
| job.queue_task() submit
| job.run_all_tasks() run
| job.yield_tasks_unordered() run
- join
- Chain.join() join
- knowns
- Building a model from
| automodel() prepare
| loopmodel() prepare
| alignment.check_structure_structure() check
- last
- actions.write_structure() write
| actions.charmm_trajectory() write
- last_scales
- schedule() create
| schedule.make_for_model() trim
- Lennard-Jones
- Frequently asked questions (FAQ)
| dope_loopmodel reference
| Residues with defined topology,
| Block (BLK) residues with
| Static and dynamic restraints
| Mathematical forms of restraints
| Rigid bodies
| energy_data.contact_shell nonbond
| energy_data.dynamic_lennard calculate
| energy_data.lennard_jones_switch Lennard-Jones
| Restraints.make() make
| selection.assess_dope() assess
| physical.values() create
| Function
| Lennard-Jones restraint
- level
- log.level() Set
- Libraries.parameters
- Libraries.parameters parameter
- Libraries.topology
- Libraries.topology topology
- library
- gbsa.Scorer() create
- library_schedule
- automodel() prepare
- local_alignment
- alignment.align() align
| alignment.salign() align
- local_slave
- local_slave() create
- local_slave()
- local_slave() create
- log object
- The log object: controlling
- log.level()
- log.level() Set
| TOP to Python correspondence
- log.minimal()
- log.minimal() display
- log.none()
- log.none() display
- log.verbose()
- Useful SALIGN information and
| log.verbose() display
- log.very_verbose()
- log.very_verbose() display
- loop modeling
- Loop optimization
| Loop modeling method
- loop_assess_methods
- loopmodel() prepare
- loopmodel
- loopmodel() prepare
- loopmodel class
- loopmodel reference
- loopmodel()
- loopmodel() prepare
| dope_loopmodel() prepare
- loopmodel.build_ini_loop()
- loopmodel.build_ini_loop() create
- loopmodel.get_loop_model_filename()
- automodel.get_model_filename() get
| loopmodel.get_loop_model_filename() get
- loopmodel.loop.assess_methods
- automodel() prepare
- loopmodel.loop.ending_model
- Automatic loop refinement after
| loopmodel.loop.ending_model last
| Loop modeling method
- loopmodel.loop.library_schedule
- loopmodel.loop.library_schedule select
- loopmodel.loop.max_var_iterations
- loopmodel.loop.max_var_iterations select
- loopmodel.loop.md_level
- loopmodel.loop.md_level control
- loopmodel.loop.outputs
- Accessing output data after
| loopmodel.loop.outputs all
- loopmodel.loop.starting_model
- Automatic loop refinement after
| loopmodel.loop.starting_model first
| Loop modeling method
- loopmodel.loop.write_defined_only
- Changes since release 9v8
| loopmodel.loop.write_defined_only only
- loopmodel.loop.write_selection_only
- loopmodel.loop.write_selection_only write
| loopmodel.loop.write_defined_only only
- loopmodel.read_potential()
- loopmodel.read_potential() read
- loopmodel.select_loop_atoms()
- Defining loop regions for
| Refining an existing PDB
| loopmodel.select_loop_atoms() select
| Loop modeling method
- loopmodel.user_after_single_loop_model()
- loopmodel.user_after_single_loop_model() analyze
- loops
- model.loops() return
- low
- Mathematical forms of restraints
- lowderiv
- Mathematical forms of restraints
- make
- automodel.make() build
| Topology.make() make
| Restraints.make() make
- make_chains
- model.make_chains() Fetch
- make_distance
- Restraints.make_distance() make
- make_for_model
- schedule.make_for_model() trim
- make_pssmdb
- environ.make_pssmdb() Create
- make_region
- model.make_region() define
- make_valid_pdb_coordinates
- model.make_valid_pdb_coordinates() make
- malign
- alignment.malign() align
- malign3d
- alignment.malign3d() align
- matrix_comparison
- Alignment of protein sequences
- matrix_file
- alignment.id_table() calculate
| alignment.compare_sequences() compare
| alignment.compare_structures() compare
- matrix_offset
- environ.make_pssmdb() Create
| alignment.align() align
| profile.scan() Compare
| sequence_db.filter() cluster
| Profile file
- matrix_offset_3d
- Features of proteins used
- max_aln_evalue
- profile.scan() Compare
| profile.build() Build
- max_atom_shift
- quasi_newton() optimize
- max_diff_res
- sequence_db.filter() cluster
- max_gap_length
- alignment.align2d() align
- max_gaps_match
- alignment.compare_sequences() compare
- max_iterations
- automodel.max_var_iterations select
| conjugate_gradients() optimize
| molecular_dynamics() optimize
- max_nonstdres
- Chain.filter() check
- max_unaligned_res
- sequence_db.filter() cluster
- max_var_iterations
- automodel.very_fast() request
- maximal_distance
- automodel.max_ca_ca_distance Distance
| Restraints.make_distance() make
- maxlength
- model.get_insertions() return
- maxs
- saxsdata.ini_saxs() Initialization
- maxslave
- sge_qsub_job() create
- md_return
- molecular_dynamics() optimize
- md_time_step
- molecular_dynamics() optimize
- mdl
- model.res_num_from() residue
| model.orient() center
| Restraints.reindex() renumber
| schedule.make_for_model() trim
| alignment.append_model() copy
| alignment.compare_sequences() compare
| complete_pdb() read
- mdl2
- selection.superpose() superpose
- mean
- Mathematical forms of restraints
- means
- Mathematical forms of restraints
- memory
- log.level() Set
- method
- Method for comparative protein
- min_atom_shift
- conjugate_gradients() optimize
- min_base_entries
- alignment.edit() edit
- min_loop_length
- alignment.segment_matching() align
- minimal
- log.minimal() display
- minimal_chain_length
- Chain.filter() check
- minimal_resolution
- Chain.filter() check
- minimal_stdres
- Chain.filter() check
- minlength
- model.get_insertions() return
- minmax_db_seq_len
- sequence_db.read() read
- missing parameters
- Frequently asked questions (FAQ)
| Residues with defined topology,
- mixflag
- saxsdata.ini_saxs() Initialization
- mode
- modfile.File() open
- model
- model() create
- model class
- The model class: handling
- model()
- model() create
- model.assess_ga341()
- automodel() prepare
| model.seq_id sequence
| model.assess_ga341() assess
| selection.assess_dope() assess
- model.assess_normalized_dope()
- automodel() prepare
| model.assess_normalized_dope() assess
| selection.assess_dope() assess
- model.atom_range()
- Feature types
| model.atoms all
| model.atom_range() return
| The selection class: handling
- model.atoms
- Feature types
| model.atoms all
| model.atom_range() return
| The selection class: handling
| The Atom class: a
- model.build()
- Residues with defined topology,
| model.remark text
| model.build_sequence() build
| model.generate_topology() generate
| model.build() build
| model.transfer_xyz() copy
| selection.only_defined() select
| Flowchart of comparative modeling
| TOP to Python correspondence
- model.build_sequence()
- model.build_sequence() build
| alignment.append_sequence() add
- model.clear_topology()
- model.clear_topology() clear
| model.generate_topology() generate
- model.color()
- model.color() color
| TOP to Python correspondence
- model.generate_topology()
- Frequently asked questions (FAQ)
| energy_data.dynamic_sphere calculate
| energy_data.dynamic_lennard calculate
| model.read() read
| model.clear_topology() clear
| model.generate_topology() generate
| model.write_psf() write
| model.patch() patch
| model.patch_ss_templates() guess
| Restraints.make() make
| selection.rotate_dihedrals() change
| Flowchart of comparative modeling
| TOP to Python correspondence
- model.get_deletions()
- model.get_insertions() return
| model.get_deletions() return
| model.loops() return
| The selection class: handling
- model.get_insertions()
- model.get_insertions() return
| model.get_deletions() return
| model.loops() return
| The selection class: handling
- model.group_restraints
- energy_data.dynamic_modeller calculate
| model.group_restraints all
- model.last_energy
- model.last_energy last
| model.write() write
- model.loops()
- model.get_insertions() return
| model.get_deletions() return
| model.loops() return
| The selection class: handling
- model.make_chains()
- model.make_chains() Fetch
| Chain.filter() check
| TOP to Python correspondence
- model.make_region()
- Changes since release 9v8
| model.make_region() define
| Atom.biso isotropic
| TOP to Python correspondence
- model.make_valid_pdb_coordinates()
- model.make_valid_pdb_coordinates() make
- model.orient()
- model.orient() center
| TOP to Python correspondence
- model.patch()
- Including disulfide bridges
| Frequently asked questions (FAQ)
| Topology.append() append
| Parameters.append() append
| model.generate_topology() generate
| model.patch() patch
| TOP to Python correspondence
- model.patch_ss()
- model.patch_ss() guess
| TOP to Python correspondence
- model.patch_ss_templates()
- Including disulfide bridges
| model.patch_ss_templates() guess
| model.patch_ss() guess
| Flowchart of comparative modeling
| TOP to Python correspondence
- model.point()
- model.point() return
| The Point class: a
- model.read()
- Residues with defined topology,
| model() create
| model.remark text
| model.read() read
| model.write() write
| model.generate_topology() generate
| model.patch_ss() guess
| model.build() build
| model.make_chains() Fetch
| Restraints.condense() remove
| selection.rotate_dihedrals() change
| Sequence.range residue
| Sequence.rfactor R
| complete_pdb() read
| Flowchart of comparative modeling
| TOP to Python correspondence
- model.remark
- model.remark text
| model.write() write
- model.rename_segments()
- Building multi-chain models with
| model.rename_segments() rename
| TOP to Python correspondence
- model.reorder_atoms()
- model.reorder_atoms() standardize
| TOP to Python correspondence
- model.res_num_from()
- model.generate_topology() generate
| model.res_num_from() residue
| TOP to Python correspondence
- model.residue_range()
- Refining only part of
| Feature types
| model.residue_range() return
| alpha() make
| strand() make
| The selection class: handling
| Sequence.residues list
- model.resolution
- model.resolution resolution
- model.restraints
- model.restraints all
| The Restraints class: static
- model.saxs_chifun()
- model.saxs_chifun() Calculate
- model.saxs_intens()
- model.saxs_intens() Calculate
- model.saxs_pr()
- model.saxs_pr() Calculate
| saxsdata.saxs_pr_read() Read
- model.seq_id
- model.seq_id sequence
| model.write() write
| model.assess_ga341() assess
- model.to_iupac()
- model() create
| model.to_iupac() standardize
| TOP to Python correspondence
- model.transfer_xyz()
- automodel.cluster() cluster
| Residues with defined topology,
| environ.dendrogram() clustering
| model.seq_id sequence
| model.generate_topology() generate
| model.build() build
| model.transfer_xyz() copy
| selection.only_defined() select
| alignment.malign3d() align
| Flowchart of comparative modeling
| TOP to Python correspondence
- model.write()
- model.seq_id sequence
| model.last_energy last
| model.read() read
| model.write() write
| model.write_data() write
| model.make_region() define
| model.color() color
| selection.write() write
| Chain.join() join
| Flowchart of comparative modeling
| TOP to Python correspondence
- model.write_data
- Residue.curvature Mainchain
- model.write_data()
- Changes since release 9v7
| energy_data.radii_factor scale
| model.write() write
| model.write_data() write
| model.make_region() define
| Atom.biso isotropic
| Atom.accessibility atomic
| Atomic solvent accessibility
| TOP to Python correspondence
- model.write_psf()
- model.write_psf() write
| actions.charmm_trajectory() write
- model_segment
- model.read() read
| Sequence.range residue
| complete_pdb() read
| Alignment file (PIR)
- modeller_path
- job() create
- modfile.default()
- modfile.default() generate
- modfile.delete()
- modfile.delete() delete
| TOP to Python correspondence
- modfile.File()
- model.write() write
| alignment.read_one() read
| alignment.write() write
| Structure.write() write
| Chain.write() write
| modfile.File() open
- modfile.inquire()
- modfile.inquire() check
| TOP to Python correspondence
- molecular_dynamics
- molecular_dynamics() optimize
- molecular_dynamics()
- molecular_dynamics() optimize
| actions.write_structure() write
| actions.charmm_trajectory() write
| Flowchart of comparative modeling
| TOP to Python correspondence
- molpdf
- selection.energy() evaluate
| selection.assess_dope() assess
| selection.assess_dopehr() assess
| conjugate_gradients() optimize
| quasi_newton() optimize
| molecular_dynamics() optimize
- mutate
- selection.mutate() mutate
- n_exceed
- selection.debug_function() test
- n_prof_iterations
- profile.build() Build
| Profile file
- n_subopt
- Sub-optimal alignments
- natomtyp
- saxsdata.ini_saxs() Initialization
- nd_spline.add_dimension
- Mathematical forms of restraints
- neighbor_cutoff
- model.write_data() write
- ngap
- Residue.add_leading_gaps() add
| Residue.add_trailing_gaps() add
| Residue.remove_leading_gaps() remove
| Residue.remove_trailing_gaps() remove
- nmesh
- saxsdata.ini_saxs() Initialization
- nodename
- sge_pe_slave() create
| ssh_slave() create
- none
- log.none() display
- nonstd_restraints
- automodel.nonstd_restraints() add
- normalize_pp_scores
- Useful SALIGN information and
- normalize_profile
- selection.energy() evaluate
- nr
- saxsdata.ini_saxs() Initialization
- nr_exp
- saxsdata.ini_saxs() Initialization
- number_of_steps
- density.grid_search() dock
- obj
- selection.add() add
- off_diagonal
- alignment.align() align
- only_atom_types
- selection.only_atom_types() select
- only_defined
- selection.only_defined() select
- only_het_residues
- selection.only_het_residues() select
- only_mainchain
- selection.only_mainchain() select
- only_no_topology
- selection.only_no_topology() select
- only_residue_types
- selection.only_residue_types() select
- only_sidechain
- selection.only_sidechain() select
- only_std_residues
- selection.only_std_residues() select
- only_water_residues
- selection.only_water_residues() select
- open
- Mathematical forms of restraints
- optimization
- conjugate gradients
- conjugate_gradients() optimize
- molecular dynamics
- molecular_dynamics() optimize
- quasi-Newton
- quasi_newton() optimize
- user-defined
- User-defined optimizers
- variable target function method
- schedule() create
- options
- sge_qsub_job() create
- orient
- model.orient() center
- OrientData
- model.orient() center
- outfile
- sequence_db.convert() convert
- output
- model.write_data() write
| Restraints.spline() approximate
| selection.energy() evaluate
| conjugate_gradients() optimize
| actions.trace() write
| alignment.compare_structures() compare
| alignment.align3d() align
| alignment.malign3d() align
| alignment.salign() align
| Useful SALIGN information and
| sequence_db.search() search
- output_alignments
- profile.scan() Compare
- output_cod_file
- sequence_db.filter() cluster
- output_grp_file
- sequence_db.filter() cluster
- output_score_file
- profile.scan() Compare
| profile.build() Build
- output_weights_file
- Useful SALIGN information and
- overhang
- alignment.edit() edit
| alignment.align() align
| Alignment of protein structures
- overhang_auto_limit
- Alignment of protein structures
- overhang_factor
- Alignment of protein structures
- parameters
- group_restraints() create
- Parameters.append()
- Libraries.parameters parameter
| Parameters.append() append
| Parameters.read() read
- Parameters.clear()
- Parameters.clear() clear
| Parameters.read() read
- Parameters.read()
- automodel() prepare
| Parameters.read() read
| TOP to Python correspondence
- patch
- model.patch() patch
- patch_default
- model.build_sequence() build
| model.generate_topology() generate
| model.patch() patch
| complete_pdb() read
- patch_ss
- model.patch_ss() guess
- patch_ss_templates
- model.patch_ss_templates() guess
- PDB files
- in alignment files
- Alignment file (PIR)
- reading
- model.read() read
- search path
- io_data.atom_files_directory search
- temperature factor
- Changes since release 9v7
| model.write() write
| model.transfer_xyz() copy
| model.write_data() write
| model.make_region() define
| model.color() color
| selection.energy() evaluate
| alignment.compare_sequences() compare
| Atom.biso isotropic
- writing
- model.write() write
- physical.values class
- physical.values() create
- physical.values()
- environ.schedule_scale energy
| strand() make
| sheet() make
| physical.values() create
- pick
- Restraints.pick() pick
- pick_hot_cutoff
- selection.hot_atoms() atoms
- PIR format
- Alignment file (PIR)
- point
- model.point() return
- Point class
- The Point class: a
- Point.select_sphere()
- The selection class: handling
| selection.select_sphere() select
| Point.select_sphere() select
| The Atom class: a
- Point.x
- Point.x x
| The Atom class: a
- pr_smooth
- saxsdata.ini_saxs() Initialization
- prf
- alignment.append_profile() add
- principal_components
- environ.principal_components() clustering
- probe_radius
- model.write_data() write
- profile
- profile() create
- profile class
- The profile class: using
- profile()
- profile() create
- profile.build()
- environ.make_pssmdb() Create
| The profile class: using
| profile.scan() Compare
| profile.build() Build
| The sequence_db class: using
| sequence_db.read() read
| sequence_db.write() write
| Profile file
| TOP to Python correspondence
- profile.read()
- environ.make_pssmdb() Create
| alignment.to_profile() convert
| profile() create
| profile.read() read
| profile.to_alignment() profile
| profile.scan() Compare
| profile.build() Build
| sequence_db.read() read
| Profile file
| TOP to Python correspondence
- profile.scan()
- Changes since release 9v7
| environ.make_pssmdb() Create
| The profile class: using
| profile() create
| profile.scan() Compare
| pssmdb() create
| pssmdb.read() read
| TOP to Python correspondence
- profile.to_alignment()
- alignment.append_profile() add
| profile.to_alignment() profile
| profile.build() Build
| TOP to Python correspondence
- profile.write()
- profile.write() write
| profile.build() Build
| TOP to Python correspondence
- profile_format
- environ.make_pssmdb() Create
| profile.scan() Compare
- profile_list_file
- environ.make_pssmdb() Create
| profile.scan() Compare
- program updates
- MODELLER updates
- protonation states
- model.read() read
- psa_integration_step
- model.write_data() write
- pseudo atoms
- Specification of pseudo atoms
- pseudo_atom.ch2
- Specification of pseudo atoms
- pseudo_atom.ch31
- Specification of pseudo atoms
- pseudo_atom.ch32
- Specification of pseudo atoms
- pseudo_atom.gravity_center
- Specification of pseudo atoms
- PSF files
- writing
- model.write_psf() write
- pssmdb
- pssmdb() create
- pssmdb()
- pssmdb() create
- pssmdb.read()
- pssmdb() create
| pssmdb.read() read
- pssmdb_name
- environ.make_pssmdb() Create
- Python
- installation
- Obtaining and installing the
- license
- Copyright notice
- used by MODELLER
- Script file
- using MODELLER as a Python module
- Running MODELLER
- q1
- Mathematical forms of restraints
- q2
- Mathematical forms of restraints
- quasi_newton
- quasi_newton() optimize
- quasi_newton()
- Rigid bodies
| quasi_newton() optimize
- queue_task
- job.queue_task() submit
- rand_seed
- environ() create
- randomize_xyz
- selection.randomize_xyz() randomize
- read
- Topology.read() read
| Parameters.read() read
| model.read() read
| profile.read() read
| pssmdb.read() read
| sequence_db.read() read
| density.read() read
- read_one
- alignment.read_one() read
- read_potential
- loopmodel.read_potential() read
- reference_atom
- selection.superpose() superpose
- reference_distance
- selection.superpose() superpose
- refine_local
- selection.superpose() superpose
- region_size
- model.make_region() define
- reindex
- Restraints.reindex() renumber
- remove_gaps
- alignment.append() read
| alignment.read_one() read
- remove_leading_gaps
- Residue.remove_leading_gaps() remove
- remove_trailing_gaps
- Residue.remove_trailing_gaps() remove
- remove_unpicked
- Restraints.remove_unpicked() remove
- rename_segments
- model.rename_segments() rename
- renumber_residues
- model.rename_segments() rename
- reorder_atoms
- model.reorder_atoms() standardize
- report
- Restraints.symmetry.report() report
- represtyp
- saxsdata.ini_saxs() Initialization
- reread
- Structure.reread() reread
- res_num_from
- model.res_num_from() residue
- Residue class
- The Residue class: a
- Residue.add_leading_gaps()
- Residue.add_leading_gaps() add
- Residue.add_trailing_gaps()
- Residue.add_trailing_gaps() add
- Residue.alpha
- Residue.alpha
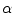
- Residue.atoms
- Residue.atoms all
- Residue.chain
- Residue.chain chain
- Residue.chi1
- Residue.chi1
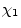
- Residue.chi2
- Residue.chi2
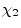
- Residue.chi3
- Residue.chi3
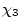
- Residue.chi4
- Residue.chi4
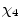
- Residue.chi5
- Residue.chi5
- Residue.code
- Changes since release 9v7
| Residue.code One-letter
- Residue.curvature
- Residue.curvature Mainchain
- Residue.get_aligned_residue()
- Residue.get_aligned_residue() get
- Residue.get_leading_gaps()
- Residue.get_leading_gaps() get
- Residue.get_trailing_gaps()
- Residue.get_trailing_gaps() get
- Residue.hetatm
- Residue.hetatm HETATM
- Residue.index
- Residue.index internal
| Residue.num PDB-style
- Residue.name
- Changes since release 9v7
| Residue.name internal
- Residue.num
- Changes since release 9v7
| Residue.num PDB-style
- Residue.omega
- Residue.omega
- Residue.pdb_name
- Changes since release 9v7
| Residue.pdb_name PDB
- Residue.phi
- Residue.phi
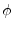 | Residue.psi
| Residue.psi 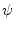 | Residue.omega
| Residue.omega 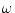 | Residue.alpha
| Residue.alpha 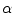 | Residue.chi1
| Residue.chi1 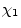 | Residue.chi2
| Residue.chi2 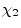 | Residue.chi3
| Residue.chi3 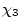 | Residue.chi4
| Residue.chi4 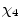 | Residue.chi5
| Residue.chi5 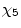 | The Dihedral class: a
| The Dihedral class: a
- Residue.psi
- Residue.psi
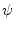
- Residue.remove_leading_gaps()
- Residue.remove_leading_gaps() remove
- Residue.remove_trailing_gaps()
- Residue.remove_trailing_gaps() remove
- residue_range
- model.residue_range() return
- residue_span_range
- energy_data.covalent_cys use
| Restraints.make() make
| Restraints.pick() pick
| conjugate_gradients() optimize
- residue_span_sign
- Restraints.make() make
- residue_type
- model.patch() patch
| selection.mutate() mutate
- residues
- Frequently asked questions (FAQ)
| model.patch() patch
| alpha() make
| strand() make
- resolution
- density.read() read
- restraint_group
- Restraints.make() make
- restraint_sel_atoms
- Restraints.make() make
| Restraints.pick() pick
- restraint_stdev
- Restraints.make_distance() make
- restraint_stdev2
- Restraints.make_distance() make
- restraint_type
- Restraints.make() make
| Restraints.unpick_redundant() unselect
- restraints
- feature types
- Feature types
- mathematical forms
- Mathematical forms of restraints
- pseudo atoms
- Specification of pseudo atoms
- rigid bodies
- Rigid bodies
- static versus dynamic
- Static and dynamic restraints
- symmetry
- Symmetry restraints
- violations
- Restraint violations
- Restraints class
- The Restraints class: static
- Restraints.add()
- Frequently asked questions (FAQ)
| Static and dynamic restraints
| Restraints.make() make
| Restraints.add() add
| Restraints.unpick() unselect
| alpha() make
| strand() make
| sheet() make
| cispeptide() creates
| TOP to Python correspondence
- Restraints.append()
- Frequently asked questions (FAQ)
| Restraints.append() read
| Restraints.write() write
| TOP to Python correspondence
- Restraints.clear()
- Restraints.pseudo_atoms all
| Restraints.clear() delete
| Restraints.append() read
- Restraints.condense()
- Frequently asked questions (FAQ)
| Restraints.unpick_redundant() unselect
| Restraints.remove_unpicked() remove
| Restraints.condense() remove
| Restraints.unpick() unselect
| Restraints.reindex() renumber
| alpha() make
| strand() make
| TOP to Python correspondence
- Restraints.excluded_pairs
- Excluded pairs
| Restraints.excluded_pairs all
- Restraints.make()
- Frequently asked questions (FAQ)
| Coordinate files and derivative
| Residues with defined topology,
| Static and dynamic restraints
| model.patch_ss_templates() guess
| Restraints.make() make
| Restraints.make_distance() make
| Restraints.pick() pick
| Restraints.spline() approximate
| Restraints.append() read
| Restraints.write() write
| Atomic solvent accessibility
| Flowchart of comparative modeling
| TOP to Python correspondence
- Restraints.make_distance()
- Frequently asked questions (FAQ)
| automodel.max_ca_ca_distance Distance
| Block (BLK) residues with
| Restraints.make() make
| Restraints.make_distance() make
| Atomic solvent accessibility
- Restraints.nonbonded_pairs
- Restraints.nonbonded_pairs all
- Restraints.pick()
- Frequently asked questions (FAQ)
| Restraints.pick() pick
| selection.hot_atoms() atoms
| schedule() create
| Flowchart of comparative modeling
| TOP to Python correspondence
- Restraints.pseudo_atoms
- Specification of pseudo atoms
| Restraints.pseudo_atoms all
- Restraints.reindex()
- Restraints.reindex() renumber
| TOP to Python correspondence
- Restraints.remove_unpicked()
- Restraints.pick() pick
| Restraints.remove_unpicked() remove
| Restraints.condense() remove
- Restraints.rigid_bodies
- Rigid bodies
| Restraints.rigid_bodies all
- Restraints.spline()
- Changes since release 9v8
| Restraints.make() make
| Restraints.spline() approximate
| User-defined restraint forms
| TOP to Python correspondence
- Restraints.symmetry
- Symmetry restraints
| Restraints.symmetry all
- Restraints.symmetry.report()
- Restraints.symmetry.report() report
- Restraints.unpick()
- Frequently asked questions (FAQ)
| Restraints.unpick() unselect
| alpha() make
| strand() make
| TOP to Python correspondence
- Restraints.unpick_all()
- Restraints.unpick_all() unselect
| Restraints.pick() pick
- Restraints.unpick_redundant()
- Restraints.make() make
| Restraints.unpick_redundant() unselect
| Restraints.condense() remove
- Restraints.write()
- Static and dynamic restraints
| Restraints.write() write
| Flowchart of comparative modeling
| TOP to Python correspondence
- restraints_filter
- Restraints.pick() pick
- restyp_lib_file
- environ() create
- rho_solv
- saxsdata.ini_saxs() Initialization
- rigid bodies
- Rigid bodies
- rigid_body
- Rigid bodies
- rms_cutoff
- selection.superpose() superpose
| Useful SALIGN information and
- rms_cutoffs
- alignment.compare_structures() compare
- root_name
- alignment.segment_matching() align
| modfile.default() generate
- rotate_dihedrals
- selection.rotate_dihedrals() change
- rotate_mass_center
- selection.rotate_mass_center() rotate
- rotate_origin
- selection.rotate_origin() rotate
- rr_file
- environ.make_pssmdb() Create
| alignment.compare_sequences() compare
| alignment.align() align
| alignment.align2d() align
| alignment.malign() align
| Features of proteins used
| Useful SALIGN information and
| alignment.segment_matching() align
| profile.scan() Compare
| sequence_db.search() search
| sequence_db.filter() cluster
| Profile file
- run_all_tasks
- job.run_all_tasks() run
- run_cmd
- slave.run_cmd() run
- s_hi
- saxsdata.ini_saxs() Initialization
- s_hybrid
- saxsdata.ini_saxs() Initialization
- s_low
- saxsdata.ini_saxs() Initialization
- s_max
- saxsdata.ini_saxs() Initialization
- s_min
- saxsdata.ini_saxs() Initialization
- salign
- alignment.salign() align
- SalignData
- alignment.salign() align
- saxs_chifun
- model.saxs_chifun() Calculate
- saxs_intens
- model.saxs_intens() Calculate
- saxs_pr
- model.saxs_pr() Calculate
- saxs_pr_read
- saxsdata.saxs_pr_read() Read
- saxs_read
- saxsdata.saxs_read() Read
- saxsdata
- saxsdata() create
- saxsdata class
- The saxsdata class: using
- saxsdata()
- saxsdata() create
- saxsdata.ini_saxs()
- saxsdata() create
| saxsdata.ini_saxs() Initialization
| saxsdata.saxs_read() Read
- saxsdata.saxs_pr_read()
- saxsdata.saxs_pr_read() Read
- saxsdata.saxs_read()
- saxsdata.saxs_read() Read
- scale_factor
- Rigid bodies
- scan
- profile.scan() Compare
- schedule
- schedule() create
- schedule class
- The schedule class: variable
- schedule()
- schedule() create
| schedule.make_for_model() trim
| schedule.write() write
- schedule.make_for_model()
- schedule() create
| schedule.make_for_model() trim
| TOP to Python correspondence
- schedule.write()
- schedule.write() write
| TOP to Python correspondence
- schedule_scale
- selection.hot_atoms() atoms
| selection.energy() evaluate
| selection.debug_function() test
| selection.assess_dope() assess
| conjugate_gradients() optimize
- score
- model.assess_ga341() assess
- score_statistics
- profile.scan() Compare
- Scorer
- gbsa.Scorer() create
- script file
- Script file
- search
- sequence_db.search() search
- search_group_list
- sequence_db.search() search
- search_randomizations
- sequence_db.search() search
- search_sort
- sequence_db.search() search
- search_top_list
- sequence_db.search() search
- segment_cutoff
- alignment.segment_matching() align
- segment_growth_c
- alignment.segment_matching() align
- segment_growth_n
- alignment.segment_matching() align
- segment_ids
- model.rename_segments() rename
- segment_matching
- alignment.segment_matching() align
- segment_report
- alignment.segment_matching() align
- segment_shift
- alignment.segment_matching() align
- select_atoms
- automodel.select_atoms() select
- select_loop_atoms
- loopmodel.select_loop_atoms() select
- select_sphere
- selection.select_sphere() select
| Point.select_sphere() select
- selection
- selection() create
| selection.hot_atoms() atoms
- selection class
- The selection class: handling
- selection()
- Refining only part of
| model.point() return
| model.get_insertions() return
| model.get_deletions() return
| model.loops() return
| The selection class: handling
| selection() create
| selection.select_sphere() select
| selection.only_atom_types() select
| selection.only_residue_types() select
| selection.translate() translate
| selection.rotate_origin() rotate
| selection.rotate_mass_center() rotate
| Point.select_sphere() select
- selection.add()
- The selection class: handling
| selection.add() add
- selection.assess_dope()
- automodel() prepare
| dope_loopmodel reference
| Rigid bodies
| model.assess_normalized_dope() assess
| selection.assess_dope() assess
| selection.assess_dopehr() assess
| Sequence.chains list
- selection.assess_dopehr()
- automodel() prepare
| dopehr_loopmodel reference
| selection.assess_dopehr() assess
- selection.by_residue()
- The selection class: handling
| selection.by_residue() make
- selection.debug_function()
- selection.debug_function() test
| physical.values() create
| TOP to Python correspondence
- selection.energy()
- Including disulfide bridges
| Frequently asked questions (FAQ)
| Mathematical forms of restraints
| model.write() write
| model.generate_topology() generate
| Restraints.nonbonded_pairs all
| selection.energy() evaluate
| selection.debug_function() test
| selection.assess_dope() assess
| Atom.biso isotropic
| complete_pdb() read
| Function
| Flowchart of comparative modeling
| TOP to Python correspondence
- selection.extend_by_residue()
- selection.extend_by_residue() extend
| selection.hot_atoms() atoms
- selection.hot_atoms()
- model.generate_topology() generate
| selection.hot_atoms() atoms
| Function
| TOP to Python correspondence
- selection.mutate()
- selection.mutate() mutate
| TOP to Python correspondence
- selection.only_atom_types()
- The selection class: handling
| selection.only_atom_types() select
- selection.only_defined()
- The selection class: handling
| selection.only_defined() select
- selection.only_het_residues()
- The selection class: handling
| selection.only_het_residues() select
- selection.only_mainchain()
- The selection class: handling
| selection.only_mainchain() select
| selection.only_sidechain() select
- selection.only_no_topology()
- The selection class: handling
| selection.only_no_topology() select
- selection.only_residue_types()
- The selection class: handling
| selection.only_residue_types() select
- selection.only_sidechain()
- The selection class: handling
| selection.only_sidechain() select
- selection.only_std_residues()
- The selection class: handling
| selection.only_std_residues() select
- selection.only_water_residues()
- The selection class: handling
| selection.only_water_residues() select
- selection.randomize_xyz()
- selection.randomize_xyz() randomize
| schedule() create
| Flowchart of comparative modeling
| TOP to Python correspondence
- selection.rotate_dihedrals()
- selection.rotate_dihedrals() change
| TOP to Python correspondence
- selection.rotate_mass_center()
- selection.rotate_origin() rotate
| selection.rotate_mass_center() rotate
- selection.rotate_origin()
- selection.rotate_origin() rotate
| selection.rotate_mass_center() rotate
| TOP to Python correspondence
- selection.select_sphere()
- The selection class: handling
| selection.select_sphere() select
| Point.select_sphere() select
- selection.superpose()
- selection.superpose() superpose
| alignment.compare_structures() compare
| TOP to Python correspondence
- selection.transform()
- selection.rotate_origin() rotate
| selection.rotate_mass_center() rotate
| selection.transform() transform
| TOP to Python correspondence
- selection.translate()
- selection.translate() translate
| TOP to Python correspondence
- selection.unbuild()
- selection.unbuild() undefine
| TOP to Python correspondence
- selection.write()
- model.write() write
| selection.write() write
- selection_segment
- Alignment file (PIR)
- send_data
- Communicator.send_data() send
- seq
- Sequence.get_num_equiv() get
| Sequence.get_sequence_identity() get
- seq_database_file
- sequence_db.read() read
| sequence_db.search() search
- seq_database_format
- sequence_db.filter() cluster
- seqid_cut
- sequence_db.filter() cluster
- sequence
- automodel() prepare
| loopmodel() prepare
| selection.rotate_dihedrals() change
- Sequence class
- The Sequence class: a
- Sequence.atom_file
- model.generate_topology() generate
| alignment.append_model() copy
| alignment.malign3d() align
| Sequence.atom_file PDB
| Structure.reread() reread
| TOP to Python correspondence
- Sequence.chains
- The model class: handling
| model.atoms all
| The selection class: handling
| Sequence.chains list
| The Chain class: a
- Sequence.code
- alignment.append_model() copy
| Sequence.code alignment
| Alignment file (PIR)
| Profile file
| TOP to Python correspondence
- Sequence.get_num_equiv()
- Sequence.get_num_equiv() get
- Sequence.get_sequence_identity()
- Sequence.get_sequence_identity() get
- Sequence.name
- Sequence.name protein
- Sequence.prottyp
- Sequence.prottyp protein
- Sequence.range
- Sequence.range residue
- Sequence.residues
- Feature types
| The model class: handling
| model.atoms all
| model.residue_range() return
| The selection class: handling
| Sequence.residues list
| The Residue class: a
- Sequence.resolution
- Sequence.resolution structure
- Sequence.rfactor
- model.read() read
| Sequence.rfactor R
- Sequence.source
- Sequence.source source
- Sequence.transfer_res_prop()
- alignment.write() write
| Sequence.transfer_res_prop() transfer
- sequence_db
- sequence_db() create
- sequence_db class
- The sequence_db class: using
- sequence_db()
- sequence_db() create
- sequence_db.convert()
- sequence_db.read() read
| sequence_db.convert() convert
- sequence_db.filter()
- Changes since release 9v7
| The sequence_db class: using
| sequence_db() create
| sequence_db.read() read
| sequence_db.filter() cluster
| TOP to Python correspondence
- sequence_db.read()
- Changes since release 9v7
| alignment.append() read
| sequence_db() create
| sequence_db.read() read
| sequence_db.convert() convert
| sequence_db.search() search
| sequence_db.filter() cluster
| Profile file
| TOP to Python correspondence
- sequence_db.search()
- The sequence_db class: using
| sequence_db.search() search
| TOP to Python correspondence
- sequence_db.write()
- sequence_db.write() write
| sequence_db.convert() convert
| TOP to Python correspondence
- sge_pe_job
- sge_pe_job() create
- sge_pe_job()
- sge_pe_job() create
- sge_pe_slave
- sge_pe_slave() create
- sge_pe_slave()
- sge_pe_slave() create
- sge_qsub_job
- sge_qsub_job() create
- sge_qsub_job()
- sge_qsub_job() create
| job.run_all_tasks() run
- sge_qsub_slave
- sge_qsub_slave() create
- sge_qsub_slave()
- sge_qsub_slave() create
- sheet
- sheet() make
- sheet()
- sheet() make
- sheet_h_bonds
- sheet() make
- signif_cutoff
- sequence_db.search() search
- similarity_flag
- Features of proteins used
| Alignment of protein sequences
| Sub-optimal alignments
| Alignments using external restraints
| Useful SALIGN information and
- skip
- actions.write_structure() write
| actions.trace() write
| actions.charmm_trajectory() write
- slave.run_cmd()
- Parallel job support
| job() create
| Communicator.send_data() send
| slave.run_cmd() run
- smoothing_window
- selection.energy() evaluate
- soft-sphere
- Frequently asked questions (FAQ)
| dope_loopmodel reference
| Block (BLK) residues with
| Static and dynamic restraints
| The energy_data class: objective
| energy_data.update_dynamic nonbond
| energy_data.sphere_stdv soft-sphere
| energy_data.dynamic_sphere calculate
| energy_data.radii_factor scale
| Restraints.make() make
| physical.values() create
| schedule() create
| Function
- solvation_model
- gbsa.Scorer() create
- spaceflag
- saxsdata.ini_saxs() Initialization
- special_patches
- automodel.special_patches() add
| model.build_sequence() build
| complete_pdb() read
- special_restraints
- automodel.special_restraints() add
- spline
- Restraints.spline() approximate
- spline_dx
- Restraints.make() make
| Restraints.spline() approximate
- spline_min_points
- Restraints.make() make
| Restraints.spline() approximate
- spline_on_site
- Restraints.make() make
- spline_range
- Restraints.make() make
- ssh_command
- ssh_slave() create
- ssh_slave
- ssh_slave() create
- ssh_slave()
- ssh_slave() create
- start
- model.atom_range() return
| model.residue_range() return
| actions.write_structure() write
| job.start() start
- start_type
- density.grid_search() dock
- statistical potential
- Loop modeling method
- stdev
- Mathematical forms of restraints
- stdevs
- Mathematical forms of restraints
- steps
- schedule() create
- strand
- strand() make
- strand()
- strand() make
| sheet() make
- Structure.reread()
- Structure.reread() reread
- Structure.write()
- Structure.write() write
- structure_types
- Chain.filter() check
- submodel
- Topology.make() make
- subopt_offset
- Sub-optimal alignments
- summary_file
- profile.scan() Compare
- superpose
- selection.superpose() superpose
- superpose_refine
- selection.superpose() superpose
- SuperposeData
- selection.superpose() superpose
- surftyp
- model.write_data() write
- swap_atoms_in_res
- selection.superpose() superpose
- symmetry
- Symmetry restraints
- symmetry restraints
- Symmetry restraints
- system
- environ.system() execute
- temperature
- molecular_dynamics() optimize
| density.grid_search() dock
- terms
- selection.energy() evaluate
- time_mark
- info.time_mark() print
- to_alignment
- profile.to_alignment() profile
- to_iupac
- model.to_iupac() standardize
- to_profile
- alignment.to_profile() convert
- Topology.append()
- Libraries.topology topology
| Topology.append() append
| Topology.read() read
| selection.rotate_dihedrals() change
| TOP to Python correspondence
- Topology.clear()
- Topology.append() append
| Topology.clear() clear
| Topology.read() read
- Topology.make()
- Topology.append() append
| Topology.make() make
| Topology.submodel select
| Topology.write() write
| TOP to Python correspondence
- Topology.read()
- automodel() prepare
| Topology.read() read
| Topology.submodel select
| TOP to Python correspondence
- Topology.submodel
- Frequently asked questions (FAQ)
| energy_data.radii_factor scale
| Topology.append() append
| Topology.make() make
| Topology.submodel select
| model.generate_topology() generate
| model.patch_ss() guess
| model.write_data() write
| TOP to Python correspondence
- Topology.write()
- Topology.write() write
| TOP to Python correspondence
- trace
- actions.trace() write
- transfer_res_num
- complete_pdb() read
- transfer_res_prop
- Sequence.transfer_res_prop() transfer
- transfer_xyz
- model.transfer_xyz() copy
- transform
- selection.transform() transform
- translate
- selection.translate() translate
- translate_type
- density.grid_search() dock
- tutorial
- Using MODELLER for comparative
- unbuild
- selection.unbuild() undefine
- unpick
- Restraints.unpick() unselect
- unpick_all
- Restraints.unpick_all() unselect
- unpick_redundant
- Restraints.unpick_redundant() unselect
- use_conv
- saxsdata.ini_saxs() Initialization
- use_lookup
- saxsdata.ini_saxs() Initialization
- use_offset
- saxsdata.ini_saxs() Initialization
- use_parallel_job
- automodel.use_parallel_job() parallelize
- use_rolloff
- saxsdata.ini_saxs() Initialization
- user_after_single_loop_model
- loopmodel.user_after_single_loop_model() analyze
- user_after_single_model
- automodel.user_after_single_model() analyze
- values
- Mathematical forms of restraints
| physical.values() create
- varatom
- alignment.compare_structures() compare
- variability_file
- alignment.compare_sequences() compare
- variable target function method
- Changing the default optimization
- verbose
- log.verbose() display
- very_fast
- automodel.very_fast() request
- very_verbose
- log.very_verbose() display
- viol_report_cut
- selection.hot_atoms() atoms
| selection.energy() evaluate
- viol_report_cut2
- selection.energy() evaluate
- virtual_atom.ch1
- Specification of pseudo atoms
- virtual_atom.ch1a
- Specification of pseudo atoms
- virtual_atom.ch2
- Specification of pseudo atoms
- voxel_size
- density.read() read
- VTFM
- see variable target function method
- weights
- Mathematical forms of restraints
- weights_type
- Features of proteins used
- write
- Topology.write() write
| model.write() write
| Restraints.write() write
| selection.write() write
| schedule.write() write
| alignment.write() write
| Structure.write() write
| Chain.write() write
| profile.write() write
| sequence_db.write() write
- write_all_atoms
- actions.write_structure() write
- write_data
- model.write_data() write
- write_fit
- alignment.malign3d() align
| Features of proteins used
| Useful SALIGN information and
- write_intermediates
- Changes since release 9v7
- write_psf
- model.write_psf() write
- write_structure
- actions.write_structure() write
- write_whole_pdb
- alignment.malign3d() align
| Useful SALIGN information and
- wswitch
- saxsdata.ini_saxs() Initialization
- yield_tasks_unordered
- job.yield_tasks_unordered() run
- Z-score
- model.assess_normalized_dope() assess
- z_comb
- model.assess_ga341() assess
- z_pair
- model.assess_ga341() assess
- z_surf
- model.assess_ga341() assess
Automatic builds
2011-03-29