- environ() -- create a new MODELLER environment
- environ.io -- default input parameters
- environ.edat -- default objective function parameters
- environ.libs -- MODELLER libraries
- environ.schedule_scale -- energy function scaling factors
- environ.dendrogram() -- clustering
- environ.principal_components() -- clustering
- environ.system() -- execute system command
- environ.make_pssmdb() -- Create a database of PSSMs given a list of profiles
- energy_data() -- create a new set of objective function parameters
- energy_data.contact_shell -- nonbond distance cutoff
- energy_data.update_dynamic -- nonbond recalculation threshold
- energy_data.sphere_stdv -- soft-sphere standard deviation
- energy_data.dynamic_sphere -- calculate soft-sphere overlap restraints
- energy_data.dynamic_lennard -- calculate Lennard-Jones restraints
- energy_data.dynamic_coulomb -- calculate Coulomb restraints
- energy_data.dynamic_modeller -- calculate non-bonded spline restraints
- energy_data.excl_local -- exclude certain local pairs of atoms
- energy_data.radii_factor -- scale atomic radii
- energy_data.lennard_jones_switch -- Lennard-Jones switching parameters
- energy_data.coulomb_switch -- Coulomb switching parameters
- energy_data.relative_dielectric -- relative dielectric
- energy_data.covalent_cys -- use disulfide bridges in residue distance
- energy_data.nonbonded_sel_atoms -- control interaction with picked atoms
- energy_data.nlogn_use -- select non-bond list generation algorithm
- energy_data.energy_terms -- user-defined global energy terms
- io_data() -- create a new input parameters object
- io_data.hetatm -- whether to read HETATM records
- io_data.hydrogen -- whether to read hydrogen atoms
- io_data.water -- whether to read water molecules
- io_data.atom_files_directory -- search path for coordinate files
- libraries.topology -- topology library information
- libraries.parameters -- parameter library information
- topology.append() -- append residue topology library
- topology.clear() -- clear residue topology library
- topology.read() -- read residue topology library
- parameters.append() -- append parameters library
- parameters.clear() -- clear parameters library
- parameters.read() -- read parameters library
- topology.make() -- make a subset topology library
- topology.submodel -- select topology model type
- topology.write() -- write residue topology library
- model() -- create a new 3D model
- model.seq_id -- sequence identity between the model and templates
- model.resolution -- resolution of protein structure
- model.last_energy -- last objective function value
- model.remark -- text remark(s)
- model.restraints -- all static restraints which act on the model
- model.group_restraints -- all restraints which act on atom groups
- model.atoms -- all atoms in the model
- model.residues -- all residues in the model
- model.chains -- all chains in the model
- model.point() -- return a point in Cartesian space
- model.atom_range() -- return a subset of all atoms
- model.residue_range() -- return a subset of all residues
- model.get_insertions() -- return a list of all insertions
- model.get_deletions() -- return a list of all deletions
- model.loops() -- return a list of all loops
- model.read() -- read coordinates for MODEL
- model.build_sequence() -- build model from a sequence of one-letter codes
- model.write() -- write MODEL
- model.clear_topology() -- clear model topology
- model.generate_topology() -- generate MODEL topology
- model.write_psf() -- write molecular topology to PSF file
- model.patch() -- patch MODEL topology
- model.patch_ss_templates() -- guess MODEL disulfides from templates
- model.patch_ss() -- guess MODEL disulfides from model structure
- model.build() -- build MODEL coordinates from topology
- model.transfer_xyz() -- copy templates' coordinates to MODEL
- model.res_num_from() -- residue numbers from MODEL2 to MODEL
- model.rename_segments() -- rename MODEL segments
- model.to_iupac() -- standardize certain dihedral angles
- model.reorder_atoms() -- standardize order of MODEL atoms
- model.orient() -- center and orient MODEL
- model.write_data() -- write derivative MODEL data
- model.make_region() -- define a random surface patch of atoms
- model.color() -- color MODEL according to alignment
- model.make_chains() -- Fetch sequences from PDB file
- model.saxs_intens() -- Calculate SAXS intensity from model
- model.saxs_pr() -- Calculate
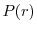 of model
of model
- model.saxs_chifun() -- Calculate SAXS score chi from model
- model.assess_ga341() -- assess a model with the GA341 method
- restraints.rigid_bodies -- all rigid bodies
- restraints.pseudo_atoms -- all pseudo atoms
- restraints.excluded_pairs -- all excluded pairs
- restraints.symmetry -- all symmetry restraints
- restraints.symmetry.report() -- report violated symmetry restraints
- restraints.make() -- make restraints
- restraints.make_distance() -- make distance restraints
- restraints.unpick_all() -- unselect all restraints
- restraints.clear() -- delete all restraints
- restraints.pick() -- pick restraints for selected atoms
- restraints.unpick_redundant() -- unselect redundant restraints
- restraints.remove_unpicked() -- remove unselected restraints
- restraints.condense() -- remove unselected or redundant restraints
- restraints.add() -- add restraint
- restraints.unpick() -- unselect restraints
- restraints.reindex() -- renumber model restraints using another model
- restraints.spline() -- approximate restraints by splines
- restraints.append() -- read spatial restraints
- restraints.write() -- write spatial restraints
- selection() -- create a new selection
- selection.add() -- add objects to selection
- selection.extend_by_residue() -- extend selection by residue
- selection.by_residue() -- make sure all residues are fully selected
- selection.select_sphere() -- select all atoms within radius
- selection.only_mainchain() -- select only mainchain atoms
- selection.only_sidechain() -- select only sidechain atoms
- selection.only_atom_types() -- select only atoms of given types
- selection.only_residue_types() -- select only atoms of given residue type
- selection.only_std_residues() -- select only standard residues
- selection.only_no_topology() -- select only residues without topology
- selection.only_het_residues() -- select only HETATM residues
- selection.only_defined() -- select only atoms with defined coordinates
- selection.write() -- write selection coordinates to a file
- selection.translate() -- translate all coordinates
- selection.rotate_origin() -- rotate coordinates about origin
- selection.rotate_mass_center() -- rotate coordinates about mass center
- selection.transform() -- transform coordinates with a matrix
- selection.mutate() -- mutate selected residues
- selection.randomize_xyz() -- randomize selected coordinates
- selection.superpose() -- superpose model on selection given alignment
- selection.rotate_dihedrals() -- change dihedral angles
- selection.unbuild() -- undefine coordinates
- selection.hot_atoms() -- atoms violating restraints
- selection.energy() -- evaluate atom selection given restraints
- selection.debug_function() -- test code self-consistency
- selection.assess_dope() -- assess a model selection with the DOPE method
- selection.assess_dopehr() -- assess a model with the DOPE-HR method
- conjugate_gradients() -- optimize atoms given restraints, with CG
- quasi_newton() -- optimize atoms with quasi-Newton minimization
- molecular_dynamics() -- optimize atoms given restraints, with MD
- actions.write_structure() -- write out the model coordinates
- actions.trace() -- write out optimization energies, etc
- actions.charmm_trajectory() -- write out a CHARMM trajectory
- User-defined optimizers
- schedule() -- create a new schedule
- schedule.make_for_model() -- trim a schedule for a model
- schedule.write() -- write optimization schedule
- group_restraints() -- create a new set of group restraints
- group_restraints.append() -- read group restraint parameters
- alignment() -- create a new alignment
- alignment.comments -- alignment file comments
- alignment.append() -- read sequences and/or their alignment
- alignment.clear() -- delete all sequences from the alignment
- alignment.check() -- check alignment for modeling
- alignment.compare_with() -- compare two alignments
- alignment.append_model() -- copy model sequence and coordinates to alignment
- alignment.append_sequence() -- add a sequence from one-letter codes
- alignment.append_profile() -- add profile sequences to the alignment
- alignment.write() -- write sequences and/or their alignment
- alignment.edit() -- edit overhangs in alignment
- alignment.describe() -- describe proteins
- alignment.id_table() -- calculate percentage sequence identities
- alignment.compare_sequences() -- compare sequences in alignment
- alignment.align() -- align two (blocks of) sequences
- alignment.align2d() -- align sequences with structures
- alignment.malign() -- align two or more sequences
- alignment.consensus() -- consensus sequence alignment
- alignment.compare_structures() -- compare 3D structures given alignment
- alignment.align3d() -- align two structures
- alignment.malign3d() -- align two or more structures
- alignment.salign() -- align two or more sequences/structures of proteins
- alignment.to_profile() -- convert alignment to profile format
- alignment.segment_matching() -- align segments
- alnsequence.code -- alignment code
- alnsequence.atom_file -- PDB file name
- alnsequence.source -- source organism
- alnsequence.name -- protein name
- alnsequence.prottyp -- protein sequence type
- alnsequence.residues -- list of residues
- alnsequence.transfer_res_prop() -- transfer residue properties
- alnsequence.get_num_equiv() -- get number of equivalences
- alnsequence.get_sequence_identity() -- get sequence identity
- chain.name -- chain ID
- chain.residues -- all residues in the chain
- chain.atoms -- all atoms in the chain
- chain.filter() -- check if this chain passes all criteria
- chain.write() -- write out chain sequence to an alignment file
- chain.atom_file_and_code() -- get suitable names for this chain
- residue.name -- internal (CHARMM) residue type name
- residue.pdb_name -- PDB (IUPAC) type name
- residue.code -- One-letter residue type code
- residue.hetatm -- HETATM indicator
- residue.index -- internal integer index
- residue.phi --
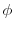 dihedral angle
dihedral angle
- residue.psi --
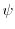 dihedral angle
dihedral angle
- residue.omega --
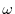 dihedral angle
dihedral angle
- residue.alpha --
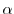 dihedral angle
dihedral angle
- residue.chi1 --
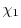 dihedral angle
dihedral angle
- residue.chi2 --
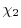 dihedral angle
dihedral angle
- residue.chi3 --
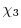 dihedral angle
dihedral angle
- residue.chi4 --
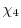 dihedral angle
dihedral angle
- residue.chi5 --
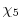 dihedral angle
dihedral angle
- dihedral.value -- current value in degrees
- dihedral.atoms -- atoms defining the angle
- dihedral.dihclass -- integer dihedral class
- atom.dvx -- objective function derivative
- atom.vx -- x component of velocity
- atom.biso -- isotropic temperature factor
- atom.occ -- occupancy
- atom.charge -- electrostatic charge
- atom.mass -- mass
- atom.name -- PDB name
- atom.residue -- residue object
- profile() -- create a new profile
- profile.read() -- read a profile of a sequence
- profile.write() -- write a profile
- profile.to_alignment() -- profile to alignment
- profile.scan() -- Compare a target profile against a database of profiles
- profile.build() -- Build a profile for a given sequence or alignment
- pssmdb() -- create a new PSSM database
- pssmdb.read() -- read a PSSM database from a file
- sequence_db() -- create a new sequence database
- sequence_db.read() -- read a database of sequences
- sequence_db.write() -- write a database of sequences
- sequence_db.convert() -- convert a database to binary format
- sequence_db.search() -- search for similar sequences
- sequence_db.filter() -- cluster sequences by sequence-identity
- density() -- create a new density map
- density.read() -- read an EM (electron microscopy) density map file
- density.grid_search() -- dock a structure into an EM (electron microscopy) density map
- saxsdata() -- create a new saxsdata structure
- saxsdata.ini_saxs() -- Initialization of SAXS data
- saxsdata.saxs_read() -- Read in SAXS data
- saxsdata.saxs_pr_read() -- Read in P(r) data
- info.version -- return the full MODELLER version number
- info.version_info -- return the version number, as a tuple
- info.build_date -- return the date this binary was built
- info.exe_type -- return the executable type of this binary
- info.debug -- returns this binary's debug flag
- info.bindir -- return MODELLER binary directory
- info.time_mark() -- print current date, time, and CPU time
- log.level() -- Set all log output levels
- log.none() -- display no log output
- log.minimal() -- display minimal log output
- log.verbose() -- display verbose log output
- log.very_verbose() -- display verbose log output, and dynamic memory information
- modfile.default() -- generate an `automatic' filename
- modfile.delete() -- delete a file
- modfile.inquire() -- check if file exists
- cispeptide() -- creates cis-peptide stereochemical restraints
- complete_pdb() -- read a PDB file, and fill in any missing residues
- job() -- create a new parallel job
- sge_pe_job() -- create a job using all Sun GridEngine (SGE) slave processes
- sge_qsub_job() -- create a job which can be expanded with Sun GridEngine 'qsub'
- job.queue_task() -- submit a task to run within the job
- job.run_all_tasks() -- run all queued tasks, and return results
- job.start() -- start all slaves for message-passing
- Communicator.send_data() -- send data
- Communicator.get_data() -- get data
- slave.run_cmd() -- run a command on the slave
- local_slave() -- create a slave running on the local machine
- sge_pe_slave() -- create a slave running on a Sun GridEngine parallel environment slave node
- sge_qsub_slave() -- create a 'qsub' slave running on a Sun GridEngine node
- ssh_slave() -- create a slave on a remote host accessed via ssh