Only one model can be calculated by this routine because the starting structure is not randomized before optimization. Only a very limited amount of the variable target function optimization with conjugate gradients is done. This is usually for a factor of 3 faster than the default procedure. For example, it takes about 17 seconds of CPU time to model a 60-residue protein on an SGI workstation with a R10000-195 processor.
# Very fast homology modelling by the MODELLER TOP routine 'model'. INCLUDE # Include the predefined TOP routines SET ALNFILE = 'alignment.ali' # alignment filename SET KNOWNS = '5fd1' # codes of the templates SET SEQUENCE = '1fdx' # code of the target SET ATOM_FILES_DIRECTORY = './:../atom_files' # directories for input atom files SET STARTING_MODEL = 2 SET ENDING_MODEL = 2 SET OUTPUT_CONTROL = 1 1 1 1 1 # SET OUTPUT = 'LONG' SET FINAL_MALIGN3D = 1 CALL ROUTINE = 'very_fast' # prepare for extremely fast optimization CALL ROUTINE = 'model' # do homology modelling
There is a pre-defined routine 'select_atoms' which selects the atoms to be moved during optimization. By default, the routine selects all atoms, but you can redefine it to select any subset of atoms and then only those atoms will be refined. They will ``feel'' the presence of other atoms via all the static and possibly dynamic restraints that include both selected and un-selected atoms. For example, the script below would refine only atoms in residues 1 and 2 (file 'examples/tutorial-model/model-segment.top'). The difference between this script and the one for loop modeling is that here the selected regions are optimized with the default optimization protocol and the default restraints, which generally include template-derived restraints. In contrast, the loop modeling routine does not use template-dependent restraints, but does a much more thorough optimization.
# Homology modelling by the MODELLER TOP routine 'model'.
# Demonstrates how to refine only a part of the model.
#
# You may want to use the more exhaustive "loop" modeling routines instead.
#
INCLUDE # Include the predefined TOP routines
SET OUTPUT_CONTROL = 1 1 1 1 0
SET ALNFILE = 'alignment.ali' # alignment filename
SET KNOWNS = '5fd1' # codes of the templates
SET SEQUENCE = '1fdx' # code of the target
SET ATOM_FILES_DIRECTORY = './:../atom_files' # directories for input atom files
SET STARTING_MODEL= 3 # index of the first model
SET ENDING_MODEL = 3 # index of the last model
# (determines how many models to calculate)
SET NONBONDED_SEL_ATOMS = 2 # selected atoms do not feel the neighbourhood
CALL ROUTINE = 'model' # do homology modelling
SUBROUTINE ROUTINE = 'select_atoms'
PICK_ATOMS SELECTION_SEGMENT='1:' '2:', SELECTION_SEARCH='segment', ;
PICK_ATOMS_SET=1, RES_TYPES='all', ATOM_TYPES='all', ;
SELECTION_FROM='all', SELECTION_STATUS='initialize'
RETURN
END_SUBROUTINE
Note that loops and insertions are already modeled by the default modeling routine, so you do not have to do anything special to get a model for the insertions. However, if you really want to focus on loops, you can use the new loop modeling routine 'loop' (Section 3.3). The selected regions are optimized independently many times by a thorough molecular dynamics/simulated annealing procedure, using sequence-dependent restraints only, no homology-derived restraints.
# Homology modelling by the MODELLER TOP routine 'model'. # Demonstrates how to refine only a part of the model. # # This can be ran with run_clustor model-loop.top, too. # # The difference with model-segment is that the loop is # refined on the basis of sequence alone, in the context # of the rest of the structure. INCLUDE # Include the predefined TOP routines SET OUTPUT_CONTROL = 1 1 1 1 1 SET SEQUENCE = '1fdx' # code of the target SET LOOP_MODEL = '1fdx.B99990001' # initial model of the target SET ATOM_FILES_DIRECTORY = './:../atom_files' # directories for input atom files # index of the first loop model: SET LOOP_STARTING_MODEL = 20 # index of the last loop model: SET LOOP_ENDING_MODEL = 23 SET LOOP_MD_LEVEL = 'refine_1' # the loop refinement method (1 fast / 3 slow) CALL ROUTINE = 'loop' # This routine picks model residues that need to be refined (necessary): SUBROUTINE ROUTINE = 'select_loop_atoms' # Uncomment if you also want to optimize the loop environment: # SET SELECTION_SEARCH = 'SPHERE_SEGMENT', SPHERE_RADIUS = 6 # 4 residue insertion (1st loop): PICK_ATOMS SELECTION_SEGMENT = '19:' '28:', SELECTION_STATUS = 'initialize' # 2 residue insertion (2nd loop): # PICK_ATOMS SELECTION_SEGMENT = '46:' '55:', SELECTION_STATUS = 'add' RETURN END_SUBROUTINE # This routine adds any special restraints (optional): # # SUBROUTINE ROUTINE = 'special_restraints' # MAKE_RESTRAINTS RESTRAINT_TYPE = 'ALPHA', RESIDUE_IDS = '46:' '55:' # RETURN # END_SUBROUTINE
This can be accomplished using the standard modeling routine. The alignment should be as follows when the chimera is a combination of proteins A and B:
proteinA aaaaaaaaaaaaaaaaaaaaaaaaaaaa---------------------------------- proteinB ----------------------------bbbbbbbbbbbbbbbbbbbbbbbbbbbbbbbbbb chimera aaaaaaaaaaaaaaaaaaaaaaaaaaaabbbbbbbbbbbbbbbbbbbbbbbbbbbbbbbbbb
In the PIR format the alignment file is:
>P1;proteinA structureX:proteinA aaaaaaaaaaaaaaaaaaaaaaaaaaaa----------------------------------* >P1;proteinB structureX:proteinB ----------------------------bbbbbbbbbbbbbbbbbbbbbbbbbbbbbbbbbb* >P1;chimera sequence:chimera aaaaaaaaaaaaaaaaaaaaaaaaaaaabbbbbbbbbbbbbbbbbbbbbbbbbbbbbbbbbb*
If no additional information is available about the relative orientation of the two domains the resulting model will probably have an incorrect relative orientation of the two domains when the overlap between A and B is non-existing or short. To obtain satisfactory relative orientation of modeled domains in such cases, orient the two template structures appropriately before the modeling.
The easiest way to achieve this is to not align that region of the template with the target sequence. If region 'bbbbbbbb' of the template should not be used as a template for region 'eeeee' of the target sequence the alignment should be like this:
template aaaaaaaaaaaaaaaaaaaaaaaa-----bbbbbbbbcccccccccccccccccccccccccccccc target ddddddddddddddddddddddddeeeee--------ffffffffffffffffffffffffffffff
The effect of this alignment is that no homology-derived restraints will be produced for region 'eeeee'.
MODELLER can restrain disulfides in two ways: automatically
(PATCH_SS_TEMPLATES or
PATCH_SS_MODEL) and
manually (PATCH).
If there is an equivalent disulfide bridge in any
of the templates aligned with the target, the PATCH_SS_TEMPLATES
command will generate appropriate disulfide bond restraints without
any other input from the user. This command is run automatically by the
'model' script used for comparative modeling. The restraints include
bond, angle and dihedral angle restraints. The SG -- SG atom pair
also becomes an excluded atom pair that is not checked for an atom-atom
overlap. The ![]() dihedral angle restraints will depend
on the conformation of the equivalent disulfides in the template
structure, as described in [Šali & Overington, 1994]. The command
PATCH_SS_MODEL is similar, except that the current structure of
MODEL, not templates, is used to guess the disulfide bonded
CYS - CYS pairs.
dihedral angle restraints will depend
on the conformation of the equivalent disulfides in the template
structure, as described in [Šali & Overington, 1994]. The command
PATCH_SS_MODEL is similar, except that the current structure of
MODEL, not templates, is used to guess the disulfide bonded
CYS - CYS pairs.
Explicit manual restraints can be added by the PATCH command relying on the PRES DISU patching residue in the CHARMM topology file. This command is used by the 'special_patches' routine that is called automatically by the 'model' script. In comparative modeling by 'model', the `manual' disulfides should be defined in the 'special_patches' routine. The PATCH command will establish the correct stereochemistry by relying on the CHARMM topology file and parameters to restrain the disulfide bond.
It is better to use PATCH_SS_TEMPLATES than PATCH where possible because the dihedral angles are restrained more precisely by using the templates than the general rules of stereochemistry.
Some CHARMM parameter files have a multiple dihedral entry for
the disulfide dihedral angle ![]() that consists of three individual
entries with periodicities of 1, 2 and 3. This is why you see three
feature restraints for a single disulfide in the output of the
ENERGY command.
that consists of three individual
entries with periodicities of 1, 2 and 3. This is why you see three
feature restraints for a single disulfide in the output of the
ENERGY command.
# This is as usual: INCLUDE SET ALIGNMENT_FILE = 'align1.ali', KNOWNS='templ1', SEQUENCE='targ1' CALL ROUTINE = 'model' STOP # Redefine the special_patches routine to include the additional disulfides # (this routine is empty by default): SUBROUTINE ROUTINE = 'special_patches' # A disulfide between residues 1 and 85 in chain A: PATCH RESIDUE_TYPE = 'DISU', RESIDUE_IDS = '1:A' '85:A' # A disulfide between residues 41 and 45 in chain B: PATCH RESIDUE_TYPE = 'DISU', RESIDUE_IDS = '41:B' '45:B' RETURN END_SUBROUTINE
MODELLER should usually be allowed to handle this automatically via the omega dihedral angle restraints, which are calculated by default.
# This is as usual:
INCLUDE
SET ALIGNMENT_FILE = 'align1.ali', KNOWNS='templ1', SEQUENCE='targ1'
CALL ROUTINE = 'model'
STOP
# Redefine the special_patches routine to force Pro to cis conformation:
# (this routine is empty by default):
SUBROUTINE ROUTINE = 'special_restraints'
CALL ROUTINE = 'cispeptide', ATOM_IDS1 = 'O:4' 'C:4' 'N:5' 'CA:5', ;
ATOM_IDS2 = 'CA:4' 'C:4' 'N:5' 'CA:5'
RETURN
END_SUBROUTINE
Restraints can be read from a file by READ_RESTRAINTS, calculated by MAKE_RESTRAINTS, or added ``manually'' by ADD_RESTRAINT. PICK_RESTRAINTS picks those restraints for objective function calculation that restrain the selected atoms only, as specified in the selected atoms set 1. Initially, all atoms are selected; this can be changed by the PICK_ATOMS command. MAKE_RESTRAINTS command for some restraint types (e.g., distance) constructs restraints of the selected type between the atoms in the selected atoms sets 2 and 3. Script 'scripts/__homcsr.top' contains examples of the PICK_ATOMS command when generating restraints by MAKE_RESTRAINTS. There are also commands for adding and deleting single restraints, ADD_RESTRAINT and DELETE_RESTRAINT, respectively. If you do CONDENSE_RESTRAINTS, the unselected restraints will be deleted. This is useful for getting rid of the unwanted restraints completely.
You can read your restraints whenever the default restraints are read.
INCLUDE SET ALIGNMENT_FILE = 'align1.ali', KNOWNS='templ1', SEQUENCE='targ1' CALL ROUTINE = 'model' STOP # Redefine the rd_restraints routine: SUBROUTINE ROUTINE = 'rd_restraints' # This is the default homology-derived restraints: READ_RESTRAINTS FILE = CSRFILE, ADD_RESTRAINTS = off # This is two additional user provided files: READ_RESTRAINTS FILE = 'my_rsrs1.rsr', ADD_RESTRAINTS = on READ_RESTRAINTS FILE = 'my_rsrs2.rsr', ADD_RESTRAINTS = on SET ADD_RESTRAINTS = off RETURN END_SUBROUTINE
This is achieved by redefining the 'special_restraints' routine, which is empty by default.
INCLUDE SET ALIGNMENT_FILE = 'align1.ali', KNOWNS='templ1', SEQUENCE='targ1' CALL ROUTINE = 'model' # Redefine the special_restraints routine: SUBROUTINE ROUTINE = 'special_restraints' # Add some restraints from a file to existing homology-derived restraints: READ_RESTRAINTS FILE = 'my_rsrs1.rsr', ADD_RESTRAINTS = on # Restrain the specified CA-CA distance to 10 angstroms (st.dev.=0.1). # Use a harmonic potential and X-Y distance group. SET ATOM_IDS 'CA:35:A' 'CA:40:A' ADD_RESTRAINT RESTRAINT_PARAMETERS = 3 1 1 27 2 2 0 10.0 0.1 SET ADD_RESTRAINTS = off RETURN END_SUBROUTINE
INCLUDE SET ALIGNMENT_FILE = 'align1.ali', KNOWNS='templ1', SEQUENCE='targ1' SET CSRFILE = 'targ1.rsr', CREATE_RESTRAINTS = 0 CALL ROUTINE = 'model'
INCLUDE SET ALIGNMENT_FILE = 'align1.ali', KNOWNS='templ1', SEQUENCE='targ1' # Specify the initial structure filename, and tell the program to read the initial file, not construct it from the templates: SET MODEL = 'targ1.ini', GENERATE_METHOD = 'read_xyz' CALL ROUTINE = 'model'
There are two different optimization approaches available within MODELLER: variable target function method (VTFM) with conjugate gradients (CG) [Šali & Blundell, 1993] and molecular dynamics (MD) with simulated annealing (SA) [Šali & Blundell, 1993]. They can both be done to a different degree (with more or less cycles of CG and MD, faster or slower schedule for VTFM and SA). The exact details are best obtained from the scripts themselves because a detailed description would probably be longer than the scripts. For example, the QUANTA and INSIGHTII implementations of MODELLER have these three levels of optimization: no optimization (only copying coordinates from templates and building the undefined atoms using internal geometry information from the RTF entries); only VTFM with CG; also MD with SA. Most of the time (70%) is spent on the MD&SA part. Our experience is that when MD&SA are used, if there are violations in the best of the 10 models, they probably come from an alignment error, not an optimizer failure (if there are no insertions longer than approximately 15 residues).
See file 'scripts/__defs.top' for the variables that could be changed and for their possible values.
INCLUDE SET ALIGNMENT_FILE = 'align1.ali', KNOWNS='templ1', SEQUENCE='targ1' # Very thorough VTFM optimization: SET LIBRARY_SCHEDULE = 1, MAX_VAR_ITERATIONS = 300 # Very thorough MD optimization: SET MD_LEVEL = 'refine1' # Repeat the whole cycle 3-times and do not stop unless obj.func. > 1E6 SET REPEAT_OPTIMIZATION = 3, MAX_MOLPDF = 1E6 CALL ROUTINE = 'model'
INCLUDE SET ALIGNMENT_FILE = 'align1.ali', KNOWNS='templ1', SEQUENCE='targ1' SET TOPOLOGY_MODEL = 1, HYDROGEN_IO = on, HETATM_IO = on, WATER_IO = on SET TOPLIB = $(LIB)/top.lib SET PARLIB = $(LIB)/par.lib CALL ROUTINE = 'model'
Water molecules are indicated by 'w' in the alignment file and the special block residue ('BLK') that does not have entries in the residue topology and parameter libraries is indicated by '.'
See Section 2.2.1 for information about block residues.
INCLUDE SET ALIGNMENT_FILE = 'align1.ali', KNOWNS='templ1', SEQUENCE='targ1' SET HETATM_IO = on, WATER_IO = on CALL ROUTINE = 'model'
The alignment file:
>P1;templ1 structureX:templ1:1::10:: FAYVI/.wwww* >P1;targ1 sequence:targ1:1::8:: -GWIV/.ww-w*
This is a painful area in all molecular modeling programs. However, CHARMM and X-PLOR provide a reasonably straightforward solution via the residue topology and parameter libraries. MODELLER uses CHARMM topology and parameter library format and also extends the options by allowing for a generic ``BLK'' residue type (Section 2.2.1). This BLK residue type circumvents the need for editing any library files, but it is not always possible to use it. Due to its conformational rigidity, it is also not as accurate as a normal residue type. In order to define a new residue type in the MODELLER libraries, you have to follow the series of steps described below. As an example, we will define the ALA residue without any hydrogen atoms. You can add an entry to the MODELLER topology or parameter file; you can also use your own topology or parameter files. For more information, please see the CHARMM manual.
- Define the new residue entry in the residue topology file (RTF),
say 'top_heav.lib'.
RESI ALA 0.00000 ATOM N NH1 -0.29792 ATOM CA CT1 0.09563 ATOM CB CT3 -0.17115 ATOM C C 0.69672 ATOM O O -0.32328 BOND CB CA N CA O C C CA C +N IMPR C CA +N O CA N C CB IC -C N CA C 1.3551 126.4900 180.0000 114.4400 1.5390 IC N CA C +N 1.4592 114.4400 180.0000 116.8400 1.3558 IC +N CA *C O 1.3558 116.8400 180.0000 122.5200 1.2297 IC CA C +N +CA 1.5390 116.8400 180.0000 126.7700 1.4613 IC N C *CA CB 1.4592 114.4400 123.2300 111.0900 1.5461 IC N CA C O 1.4300 107.0000 0.0000 122.5200 1.2297 PATC FIRS NTER LAST CTER
You can obtain an initial approximation to this entry by defining the new residue type using the residue type editor in QUANTA and then writing it to a file.
The RESI record specifies the CHARMM residue name, which can be up to four characters long and is usually the same as the PDB residue name (exceptions are the potentially charged residues where the different charge states correspond to different CHARMM residue types). The number gives the total residue charge.
The ATOM records specify the IUPAC (i.e., PDB) atom names and the CHARMM atom types for all the atoms in the residue.1.1 The number at the end of each ATOM record gives the partial atomic charge.
The BOND records specify all the covalent bonds between the atoms in the residue (e.g., there are bonds CB-CA, N-CA, O-C, etc.). In addition, symbol '+' is used to indicate the bonds to the subsequent residue in the chain (e.g., C - +N). The covalent angles and dihedral angles are calculated automatically from the list of chemical bonds.
The IMPR records specify the improper dihedral angles, generally used to restrain the planarity of various groups (e.g., peptide bonds and sidechain rings). See also below.
The IC (internal coordinate) records are used for constructing the initial Cartesian coordinates of a residue. An entry

specifies distances
 , angles
, angles 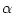 , and either dihedral angles
or improper dihedral angles
, and either dihedral angles
or improper dihedral angles  between atoms
between atoms 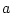 ,
,  ,
,  and
and
 , given by their IUPAC names. The improper dihedral angle
is specified when the third atom,
, given by their IUPAC names. The improper dihedral angle
is specified when the third atom,  , is preceded by a star,
'*'. As before, the '-' and '+' pre-fixes for the atom names select
the corresponding atom from the preceding and subsequent residues,
respectively. The distances are in angstroms, angles in degrees.
The distinction between the dihedral angles and
improper dihedral angles is unfortunate since they are the
same mathematically, except that by convention when using the
equations, the order of the atoms for a dihedral angle is
, is preceded by a star,
'*'. As before, the '-' and '+' pre-fixes for the atom names select
the corresponding atom from the preceding and subsequent residues,
respectively. The distances are in angstroms, angles in degrees.
The distinction between the dihedral angles and
improper dihedral angles is unfortunate since they are the
same mathematically, except that by convention when using the
equations, the order of the atoms for a dihedral angle is  and for an improper dihedral angle it is
and for an improper dihedral angle it is  .
.
The PATC record specifies the default patching residue type when the current residue type is the first or the last residue in a chain.
- You have to make sure that all the CHARMM atom types of the
new residue type occur in the MASS records at the beginning
of the topology library: Add your entry at the end of the MASS list if
necessary. If you added any new CHARMM atom types, you also have to
add them to the radii libraries, 'modlib/radii.lib' and
'modlib/radii14.lib'. These libraries list the atomic radii for the
different topology models, for the long range and 1-4 non-bonded
soft-sphere terms, respectively. The full names of the files that are
used during calculation are given by the environment variables
$RADII_LIB and $RADII14_LIB.
- Optionally, you can add the residue entry to the library of
MODELLER topology models, 'modlib/models.lib'. The runtime
version of this library is specified by the environment variable
$MODELS_LIB. This library specifies which subsets of atoms
in the residue are used for each of the possible topologies.
Currently, there are 9 topologies selected by MODEL_TOPOLOGY
(3 is default):
1 ALLH all atoms 2 POL polar hydrogens only 3 HEAV non-hydrogen atoms only 4 MCCB non-hydrogen mainchain (N, C, CA, O) and CB atoms 5 MNCH non-hydrogen mainchain atoms only 6 MCWO non-hydrogen mainchain atoms without carbonyl O 7 CA CA atoms only 8 MNSS non-hydrogen mainchain atoms and disulfide bonds 9 CA3H reduced model with a small number of sidechain interaction centers The Ala entry is:
# ALLH POLH HEAV MCCB MNCH MCWO CA MNSS CA3H * RESI ALA ATOM NH1 NH1 NH1 NH1 NH1 NH1 #### NH1 #### ATOM H HN #### #### #### #### #### #### #### ATOM CT1 CT1 CT1 CT1 CT1 CT1 CT1 CT1 CAH ATOM HB #### #### #### #### #### #### #### CH3E ATOM CT3 CT3 CT3 CT3 #### #### #### #### #### ATOM HA #### #### #### #### #### #### #### #### ATOM HA #### #### #### #### #### #### #### #### ATOM HA #### #### #### #### #### #### #### #### ATOM C C C C C C #### C #### ATOM O O O O O #### #### O ####The residue entries in this library are separated by stars. The '####' string indicates a missing atom. The atom names for the present atoms are arbitrary. The order of the atoms must be the same as in the CHARMM residue topology library. If a residue type does not have an entry in this library, all atoms are used for all topologies.
- You have to add the new residue type to the residue type library,
'modlib/restyp.lib'. The execution version of this file is
specified by the environment variable $RESTYP_LIB. For the
ALA residue,
1 | ALA | A | ALA | alanine
You would generally add the new residue type at the end of the file. There are 5 fields in each line, separated by the '|' characters. The first field is an integer index corresponding to the integer residue type. The standard residue types have their indices smaller than 24. These are also the indices corresponding to the residue-residue substitution matrices. The second field contains the list of equivalent PDB or IUPAC 3-character residue names, used in the PDB files. A list rather than a single name is allowed because PDB can unfortunately use different names for the same residue type (e.g., water can be HOH, WAT, etc.). The third field gives a single character code for the residue type, which is used in the alignment file. This does not have to be unique, but if it is not unique you cannot use it in the alignment file. Any ASCII character is fine, it does not have to be a letter. If you run out of characters you can re-define the existing ones that you do not need. The fourth field gives the four-character CHARMM residue name, as specified in the RESI record of the topology library. The last field contains an optional comment.
Every residue in the CHARMM topology file has to have an entry in the $RESTYP_LIB library, but not every residue entry in the $RESTYP_LIB library needs an entry in the residue topology file.
When you are adding a new residue type, you have to hope that the maximal number of residue types is not over-reached. If it is, a fatal error is reported at the beginning of the execution. To solve this problem, you could delete some of the un-needed existing residue types in the $RESTYP_LIB file, rather than re-compile the program with larger array sizes. You can also read your own residue type library by the READ_RESTYP_LIB command.
- In general, when you add a new residue type, you also add new
chemical bonds, angles, dihedral angles, improper dihedral angles,
and non-bonded interactions,
new in the sense that a unique combination of CHARMM atoms types
is involved whose interaction parameters are not yet specified in the
parameter library (see also Section 2.2.1).
In such a case, you will get a number of
warning and/or error messages when you generate the stereochemical
restraints by the MAKE_RESTRAINTS command. These messages
can sometimes be ignored because MODELLER will guess the
values for the missing parameters from the current Cartesian coordinates
of the model. When this is not accurate enough or if the necessary
coordinates are undefined
you have to specify the parameters explicitly in the parameter
library. Search for BOND, ANGL, DIHE, and IMPR sections in the
parameters library file and use the existing entries to guess your
new entries. Note that you can use dummy atom types 'X' to create
general dihedral (i.e., X A A X) and improper dihedral angle (i.e.,
A X X A) entries, where A stands for any of the real CHARMM
atom types.
For the dihedral angle cosine terms, the CHARMM convention for the phase
is different for 180
 from MODELLER's (Eq. 5.56).
If you use non-bonded Lennard-Jones terms, you also have to add a NONB
entry for each new atom type. If you use the default soft-sphere
non-bonded restraints, you have already taken care of it by
adding the new atom types to the $RADII_LIB and $RADII_LIB
libraries.
from MODELLER's (Eq. 5.56).
If you use non-bonded Lennard-Jones terms, you also have to add a NONB
entry for each new atom type. If you use the default soft-sphere
non-bonded restraints, you have already taken care of it by
adding the new atom types to the $RADII_LIB and $RADII_LIB
libraries.
This is even messier than defining a new residue type. As an example, we will define the patching residue for establishing a disulfide bond between two CYS residues.
PRES DISU -0.36 ! Patch for disulfides. Patch must be 1-CYS and 2-CYS. ATOM 1CB CT2 -0.10 ! ATOM 1SG SM -0.08 ! 2SG--2CB-- ATOM 2SG SM -0.08 ! / ATOM 2CB CT2 -0.10 ! -1CB--1SG DELETE ATOM 1HG1 DELETE ATOM 2HG1 BOND 1SG 2SG IC 1CA 1CB 1SG 2SG 0.0000 0.0000 180.0000 0.0000 0.0000 IC 1CB 1SG 2SG 2CB 0.0000 0.0000 90.0000 0.0000 0.0000 IC 1SG 2SG 2CB 2CA 0.0000 0.0000 180.0000 0.0000 0.0000
The PRES record specifies the CHARMM patching residue type (up to four characters). As for the normal RESI residue types, patching residue types also have to be defined in the residue type library, 'modlib/restyp.lib'.
The ATOM records have the same meaning as for the RESI residue types described above. The extension is that the IUPAC atom names (listed first) must be pre-fixed by the index of the residue that is patched. In this example, there are two CYS residues that are patched, thus the prefixes 1 and 2. When using the PATCH command, the order of the patched residues specified by RESIDUE_IDS must correspond to these indices (this is only important when the patch is not symmetric, unlike the 'DISU' patch in this example).
DELETE records specify the atoms to be deleted, the two hydrogens bonded to the two sulphurs in this case.
The BOND and IC (internal coordinate) records are the same as those for the RESI residues, except that the atom names are prefixed with the patched residue indices.
Yes. There are 'ALPHA', 'STRAND' and 'SHEET' restraint types that the MAKE_RESTRAINTS command can generate. One specifies the segment which is then restrained to the specified secondary structure conformation. For example,
# This is as usual: INCLUDE SET ALIGNMENT_FILE = 'align1.ali', KNOWNS='templ1', SEQUENCE='targ1' CALL ROUTINE = 'model' STOP # Redefine the special_restraints routine to include the secondary # structure restraints (this routine is empty by default): SUBROUTINE ROUTINE = 'special_restraints' SET ADD_RESTRAINTS = on # An alpha-helix: MAKE_RESTRAINTS RESTRAINT_TYPE = 'alpha', RESIDUE_IDS = '20' '30' # SET KEEP_DUPL_RESTR = 'new' # Two strands: MAKE_RESTRAINTS RESTRAINT_TYPE = 'STRAND', RESIDUE_IDS = '1' '6' MAKE_RESTRAINTS RESTRAINT_TYPE = 'STRAND', RESIDUE_IDS = '9' '14' # An anti-parallel sheet: MAKE_RESTRAINTS RESTRAINT_TYPE = 'SHEET', ATOM_IDS = 'N:1' 'O:14', SHEET_H-BONDS = -5 RETURN END_SUBROUTINE
This is probably because the N-terminus is patched by default with
the NTER patching residue (corresponding to -NH3![]() ) and a patched
residue must not be patched again. The solution is to turn the default
patching off by SET PATCH_DEFAULT = off before the
GENERATE_TOPOLOGY command is called.
) and a patched
residue must not be patched again. The solution is to turn the default
patching off by SET PATCH_DEFAULT = off before the
GENERATE_TOPOLOGY command is called.
Yes. You do not have to do anything special.
First, check for the error messages by searching for string '_E>''. These messages can only rarely be ignored. Next, check for the warning messages by searching for string '_W>''. These messages can almost always be ignored. If everything is OK so far, the most important part of the log file is the output of the ENERGY command for each model. This is where the violations of restraints are listed. When there are too many too violated restraints, more optimization or a different alignment is needed. What is too many and too much? It depends on the restraint type and is best learned by doing ENERGY on an X-ray structure or a good model to get a feel for it. You may also want to look at the output of command CHECK_ALIGNMENT, which should be self-explanatory. I usually ignore the other parts of the log file.
The best way to prevent knots is to start with a starting structure that is as close to the desired final model as possible. Other than that, the only solution at this point is to calculate independently many models and hope that in some runs there won't be knots. Knots usually occur when one or more neighboring long insertions (i.e., longer than 15 residues) are modeled from scratch. The reason is that an insertion is build from a randomized distorted structure that is located approximately between the two anchoring regions. Under such conditions, it is easy for the optimizer to ``fall'' into a knot and then not be able to recover from it. Sometimes knots result from an incorrect alignment, especially when more than one template is used. When the alignment is correct, knots are a result of optimization not being good enough. However, making optimization more thorough by increasing the CPU time would not be worth it on the average as knots occur relatively infrequently. The excluded volume restraints are already included in the standard comparative modeling routine.
The executable is not recognized as such on your system. Make sure you FTP the file in the binary format. Make sure the system version matches the self-descriptive name of the binary file. Also it could be related to automatic processing of files by some Web browsers. Make sure you got a binary, not the file compressed by "compress" or "gzip" command. If you are not sure about the version of your system use the most generic executable which has been compiled for lower version of operating system.
Usually more than that (dozens if you want just to detect reliable similarity, and even more if you want a real model). It is good to have at least 35-40Sometimes even 30