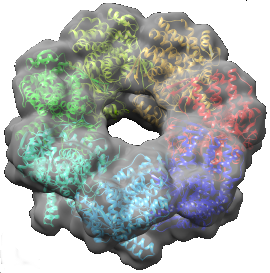
These scripts demonstrate the use of IMP, MODELLER and Chimera in the modeling of the bacterial molecular chaperone GroEL. First, MODELLER is used to generate structures for the individual components in the GroEL complex. Then, IMP is used to fit these components together into the electron microscopy density map of the entire complex.
A full description of the scripts can be found in Macromolecular assembly structures by comparative modeling and electron microscopy.
First, make a directory for the output by running mkdir output. Output
files that were generated when these scripts were run for the first time are
also provided, in the precalculate_results directory. Then, the scripts can
be run in order:
- Template identification:
scripts/script1_build_profile.py - Template(s) selection by sequence:
scripts/script2_compare_templates.py - Density map segmentation:
scripts/script3_density_segmentation.py - Template selection by fitting to a density map:
scripts/script4_score_templates_by_cc.py - Template alignment:
scripts/script5_template_alignment.py - Model building and assessment:
scripts/script6_model_building_and_assessment.pyandscripts/script7_pairwise_rmsd.py - Multiple fitting into a density map:
scripts/script8_split_density.pyandscripts/script9_symmetric_multiple_fitting.py
Author(s): Keren Lasker
Version: 1.0
License: LGPL. This library is free software; you can redistribute it and/or modify it under the terms of the GNU Lesser General Public License as published by the Free Software Foundation; either version 2 of the License, or (at your option) any later version.
Testable: Yes.
Parallelizeable: No
Publications:
- Keren Lasker, Javier A. Velazquez-Muriel, Benjamin M. Webb, Zheng Yang, Thomas E. Ferrin, Andrej Sali, Macromolecular assembly structures by comparative modeling and electron microscopy, Methods in Molecular Biology, 2011.



 Download files
Download files Verified to work with the
Verified to work with the  To install the software needed to reproduce this system with the
To install the software needed to reproduce this system with the
 To set up the environment on the UCSF Wynton cluster to run
this system, run:
To set up the environment on the UCSF Wynton cluster to run
this system, run: