This repository contains the modeling files and the analysis related to the article "Molecular Architecture of the Major Membrane Ring Component of the Nuclear Pore Complex" by Upla et al. in Structure 2017. The scripts work with IMP (version 2.6 or later).
-
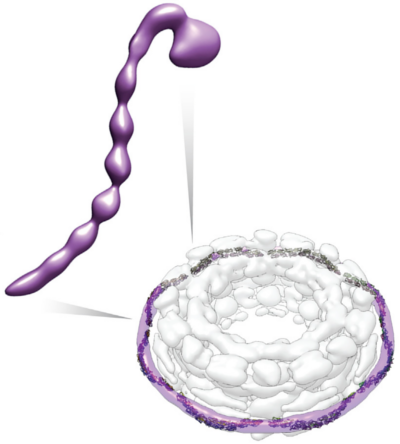
datacontains all relevant data*.pdb: Representation PDB for individual domains*.csv: Cross-links data for further validationFASTA_*.txt: Sequence filesdata/Cadherin: sequence and structure alignments between Cadherin and Pom152data/em2d_T1_truncation_1-1135: EM 2D class averages of the Pom152 1-1135 (truncated)data/em2d_T2_truncation_1-936: EM 2D class averages of the Pom152 1-936 (truncated)data/em2d_WT: EM 2D class averages of the full length Pom152data/em3d_density: EM 3D density map of the full length Pom152data/HHPred: Template search results using HHPreddata/MODELLER_all: Comparative models generated using MODELLERdata/Pom152_comparative_models_by_SAXS: Best-scoring comparative models filtered by the corresponding SAXS data -
templatecontains modeling scriptsmodeling_pdb375_482.py: modeling script -
resultscontains resulting structures and output filesresults/EM2D_selected: Validation of the Pom152 structure using selected EM 2D class averages (using multiple resolutions ranging 35 - 80 Å)results/Pom152_em3d_Final: Best-scoring Pom152 structures (clusters 0 and 1) and localization probability density mapsresults/Pom152_em3d_with_Rotational_Restraint: Refined Pom152 structure using a dihedral restraint (holding a 90 degree rotation between neighboring Ig-fold domains) -
prefilterSelection of 500 good-scoring models ranked by the combined total score. Also score log files (*.log) are included -
analysiscontains Clustering scripts and results
First, comparative models were built for each domain of Pom152:
cd data/MODELLER_all(cd 375-482 && python all_sjkim_final.py > all_sjkim_final.log): Residues 375-482(cd 516-611 && python all_sjkim_final.py > all_sjkim_final.log): Residues 516-611(cd MODELLER_27005 && python all_sjkim_final.py > all_sjkim_final.log): Residues 603-828(cd MODELLER_26996 && python all_sjkim_final.py > all_sjkim_final.log): Residues 718-1156(cd 1146-1237 && python all_sjkim_final.py > all_sjkim_final.log): Residues 1146-1237(cd 1238-1337 && python all_sjkim_final.py > all_sjkim_final.log): Residues 1238-1337
Next, integrative modeling was carried out, taking the comparative models and experimental data as input, followed by clustering and analysis:
(cd template && python modeling_pdb375_482.py)(cd analysis && python clustering.py && python precision_rmsf.py -dir kmeans_1000_2)
Author(s): Seung Joong Kim
Date: March 2017
License: CC-BY-SA-4.0. This work is freely available under the terms of the Creative Commons Attribution-ShareAlike 4.0 International License.
Publications:
- Upla P, Kim SJ, Sampathkumar P, Dutta K, Cahill SM, Chemmama IE, Williams R, Bonanno JB, Rice WJ, Stokes DL, Cowburn D, Almo SC, Sali A, Rout MP, Fernandez-Martinez J. Molecular Architecture of the Major Membrane Ring Component of the Nuclear Pore Complex, Structure, 2017, 10.1016/j.str.2017.01.006.



 Download files
Download files Verified to work with the
Verified to work with the  To install the software needed to reproduce this system with the
To install the software needed to reproduce this system with the
 To set up the environment on the UCSF Wynton cluster to run
this system, run:
To set up the environment on the UCSF Wynton cluster to run
this system, run: