Tutorial
Difficult example:
Modeling the sequence of a SARS protein. The case of the nsp16 domain from
pp1ab polyprotein.
All input and output files for this example are available to download, in either zip format (for Windows) or .tar.gz format (for Unix/Linux).
The latest outbreak of the severe acute respiratory syndrome (SARS) epidemic has led to thousands of potentially lethally infected patients and hundreds of deaths. Meanwhile, the SARS coronavirus identified as the pathogen responsible for the disaster has been isolated, and its genome sequenced. In this exercise we will try to model the sequence of the nsp16 protein of the pp1ab polyprotein from SARS. Let's first download the sequence of nsp16 defined in NCBI as a putative ribose 2'-O-methyltransferase (gi number 30133975).
>gi|30133975|ref|NP_828873.2| nsp16-pp1ab (2'-o-MT); putative ribose 2'-O-methyltransferase [SARS coronavirus] ASQAWQPGVAMPNLYKMQRMLLEKCDLQNYGENAVIPKGIMMNVAKYTQLCQYLNTLTLAVPYNMRVIHF GAGSDKGVAPGTAVLRQWLPTGTLLVDSDLNDFVSDADSTLIGDCATVHTANKWDLIISDMYDPRTKHVT KENDSKEGFFTYLCGFIKQKLALGGSIAVKITEHSWNADLYKLMGHFSWWTAFVTNVNASSSEAFLIGAN YLGKPKEQIDGYTMHANYIFWRNTNPIQLSSYSLFDMSKFPLKLRGTAVMSLKENQINDMIYSLLEKGRL IIRENNRVVVSSDILVNN
File: 30133975.faa
A template search with the BLAST and PSI-BLAST programs did not find any suitable known three-dimensional structure homologous to the nsp16 sequence. However, from the PSI-BLAST output we can conclude that the protein is closely related to RNA-directed RNA polymerases.
gi|26008094|ref|NP_742142.1| coronavirus nsp13 [Bovine coronavirus] 404 e-111 gi|37999876|sp|Q9PYA3|R1AB_CVM2 Replicase polyprotein 1ab (pp1ab... 401 e-110 gi|26007546|ref|NP_068668.2| ORF1ab polyprotein [Murine hepatiti... 401 e-110 gi|37999877|sp|P16342|R1AB_CVMA5 Replicase polyprotein 1ab (pp1a... 401 e-110 gi|7769342|gb|AAF69332.1| RNA-directed RNA polymerase [murine he... 400 e-110 gi|6625761|gb|AAF19384.1| RNA-directed RNA polymerase [murine he... 400 e-110 gi|37999878|sp|P19751|R1AB_CVMJH Replicase polyprotein 1ab (pp1a... 399 e-110 gi|93916|pir||S15760 genome polyprotein - murine hepatitis virus... 399 e-110 gi|7769353|gb|AAF69342.1| RNA-directed RNA polymerase [murine he... 399 e-110 gi|4377413|emb|CAA36202.1| unnamed protein product [Murine hepat... 399 e-110 gi|2641128|gb|AAB86818.1| RNA-directed RNA polymerase [murine he... 399 e-110 gi|7583321|gb|AAA46458.2| open reading frame 1b [murine hepatiti... 397 e-109 gi|74827|pir||VFIHJH genome polyprotein 1b - murine hepatitis vi... 397 e-109 gi|25121573|ref|NP_740620.1| coronavirus nsp13 [Murine hepatitis... 387 e-106 gi|45655908|ref|YP_003766.1| replicase polyprotein 1ab [Human Co... 367 e-100 gi|46369871|gb|AAS89765.1| ORF 1ab [Human group 1 coronavirus as... 365 e-100 gi|37999893|sp|Q9IW06|R1AB_CVPPU Replicase polyprotein 1ab (pp1a... 355 8e-97 gi|9635157|ref|NP_058422.1| replicase [Transmissible gastroenter... 355 8e-97 gi|32454345|gb|AAP82967.1| orf1ab polyprotein [SARS coronavirus ... 349 3e-95
Extracts from file: 30133975.pbo
Next the sequence from the SARS virus was submitted to the mGenThreader server for fold assignment. The server returned only one significant hit (as submitted on February 2004):
| Conf. | Net Score | E-value | PairE | SolvE | Aln Score | Aln Len | Str Len | Seq Len | Alignment | SCOP Codes |
|---|---|---|---|---|---|---|---|---|---|---|
| CERT | 0.903 | 1e-04 | -516.4 | -0.7 | 232.0 | 166 | 180 | 298 | 1ej0A0 | c.66.1.2 |
| MEDIUM | 0.650 | 0.02 | -512.7 | 1.7 | 114.0 | 151 | 173 | 298 | 1j4fA0 | - |
| MEDIUM | 0.645 | 0.022 | -502.6 | -2.7 | 122.0 | 155 | 230 | 298 | 1fbnA0 | c.66.1.3 |
| MEDIUM | 0.640 | 0.024 | -467.5 | -3.9 | 121.0 | 152 | 194 | 298 | 1dusA0 | c.66.1.4 |
| MEDIUM | 0.620 | 0.038 | -435.7 | -2.6 | 120.0 | 159 | 264 | 298 | 1i9gA0 | c.66.1.13 |
| MEDIUM | 0.606 | 0.05 | -485.2 | -1.6 | 115.0 | 166 | 186 | 298 | 1kxzA0 | c.66.1.22 |
Extracts from mGenThreader results. File: 30133975_mGenThreader.html
Alignment between the nsp16 sequence and the 1ej0A from mGenThreader results.
C; mGenThreader alignment of 30133975 and 1ej0A
C; CERT significance with an e-value of 1e-04
C; Percentage Identity = 14.4%
>P1;1ej0A
structureX:1ej0: 40 :A: 209 :A::::
-------GLRSRAWFKL----------------------------------DEIQQSDKLFKPGMTVVDL
GA------APGGWSQYVVTQIGGKGRIIACDLLPMDPIVGVDFLQGDFRDELVMKALLERVGDSKVQVVM
SDMAPNMSGTPAVDIPRAMYLVELALEMCRDVLAPGGSFVVKVFQGEGFDEYLREIRSLFTKVKVRKPDS
SRARSREVYIVATGRKP*
>P1;30133975
sequence:::::::::
ASQAWQPGVAMPNLYKMQRMLLEKCDLQNYGENAVIPKGIMMNVAKYTQLCQYLNTLTLAVPYNMRVIHF
GAGSDKGVAPG--TAVLRQWLPTGTLLVDSDLNDFVSDADSTLIGDCATVH----------TANKWDLII
SDMYDPRTKHVTKENDSKEGFFTYLCGFIKQKLALGGSIAVKITEHS-WNADLYKLMGHFSWWTAFVTNV
NA-SSSEAFLIGANYLG*
File 30133975_1ej0A_mGenThreader.ali. Red residues were manually removed from the alignment.
Five models were built for the nsp16 sequence based on the mGenThreader alignment. The file model.py shows the script used.
from modeller import *
from modeller.automodel import *
env = Environ()
a = AutoModel(env, alnfile='30133975_1ej0A_mGenThreader.ali',
knowns='1ej0A', sequence='30133975')
a.starting_model = 1
a.ending_model = 5
a.make()
File: model.py
All 5 models were then evaluated with the DOPE potential in the MODELLER program and the model 30133975.B99990001 was selected as the final model with a global score of -17031.0.
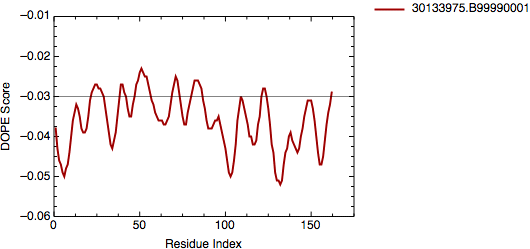
DOPE score for model 30133975.B99990001.pdb
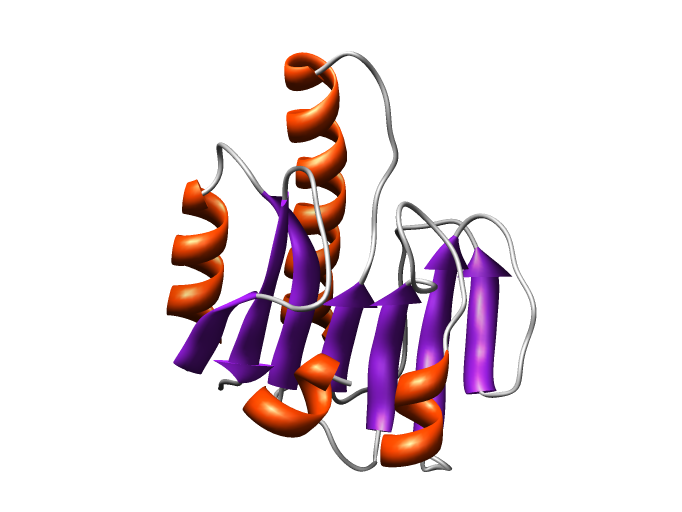
Figure of the model 30133975_1 rendered with Chimera
The PDB structure 1ej0A corresponds to a mRNA cap methylation. These proteins are found indispensable for efficient replication of many viruses and represents an active area for drug development. Nevertheless, direct inhibitors of the nsp13 enzyme may fail to suppress viral replication, as the cap-1 formation seems to be less critical than the preceding cap-0 (mGpppN) formation. The existence of the cap-1-forming enzyme in the genome would suggest that the virus also requires the AdoMet-dependent cap-0 methyltransferase. Both functions can be inhibited by carbocyclic analogs of adenosine, such as Neplanocin A or 3-deazaneplanocin A, which interfere with the AdoMet-AdoHcy metabolism of the host cell. Those compounds could complement other therapeutic strategies aimed at blocking enzymatic functions such as the RNA-dependent RNA polymerase, the protease, or the helicase encoded by the SARS virus.
This exercise was inspired by the work of Grotthuss, Wyrwicz and
Rychlewski
Letter to the Editor
"mRNA Cap-1 Methyltransferase in the SARS Genome"
Marcin von Grotthuss, Lucjan S. Wyrwicz, and Leszek Rychlewski Cell, Vol 113,
701-702, 13 June 2003