These scripts demonstrate the use of IMP, MODELLER, and PMI in the modeling of the SEA complex using 188 DSS chemical cross-links and 23 composites from affinity purification.
First, MODELLER is used to generate initial structures for the individual components where reliable templates are available. Then, IMP / PMI are used to model these components using the DSS crosslinks and the affinity purification data for the entire SEA complex.
The scripts work with the 65734ec version (develop branch) of IMP and the 47dafcc version (develop branch) of PMI.
A full description of the scripts can be found in Molecular architecture and function of the SEA complex, a modulator of the TORC1 pathway.
-
pdbcontains all input crystal structures that were deposited in PDB. -
scripts-
sj_SEA_multi_layers.pyThe main modeling script with 1:3 stoichiometry and rotational symmetry -
MODELLER/Npr2MODELLER scripts and output comparative models of Npr2 -
MODELLER/Npr3MODELLER scripts and output comparative models of Npr3 -
MODELLER/SEA1MODELLER scripts and output comparative models of SEA1 -
MODELLER/SEA2MODELLER scripts and output comparative models of SEA2 -
MODELLER/SEA3MODELLER scripts and output comparative models of SEA3 -
MODELLER/SEA4MODELLER scripts and output comparative models of SEA4
-
-
output/three_sym_cluster-
Largest output cluster as a set of RMF files. Each file is named
XX_REFINED_models_YY.rmfwhereXXidentifies the run from which the model was taken andYYthe frame number. For the publication, 885 runs were carried out. -
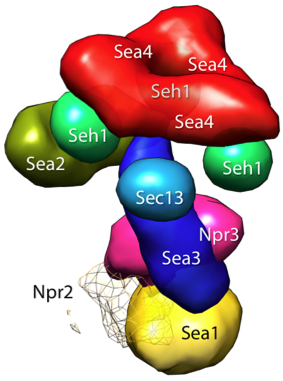
globalLocalization densities in MRC format and a Chimera session file (Chimera_three_sym.py) to display them.
-
cd scripts/MODELLER/Npr2 && python all_sjkim_final1.py: Npr2 9-127cd scripts/MODELLER/Npr2 && python all_sjkim_final2.py: Npr2 257-327cd scripts/MODELLER/Npr2 && python all_sjkim_final3.py: Npr2 563-610cd scripts/MODELLER/Npr3 && python all_sjkim_final1.py: Npr3 322-438cd scripts/MODELLER/Npr3 && python all_sjkim_final2.py: Npr3 531-577cd scripts/MODELLER/Npr3 && python all_sjkim_final3.py: Npr3 1-31cd scripts/MODELLER/Npr3 && python all_sjkim_final4.py: Npr3 950-988cd scripts/MODELLER/Npr3 && python all_sjkim_final5.py: Npr3 1083-1140cd scripts/MODELLER/SEA1 && python all_sjkim_final1.py: SEA1 101-275cd scripts/MODELLER/SEA1 && python all_sjkim_final2.py: SEA1 279-473cd scripts/MODELLER/SEA1 && python all_sjkim_final3.py: SEA1 1178-1273cd scripts/MODELLER/SEA2 && python all_sjkim_final1.py: SEA2 127-520cd scripts/MODELLER/SEA2 && python all_sjkim_final2.py: SEA2 1280-1341cd scripts/MODELLER/SEA3 && python all_sjkim_final1.py: SEA3 54-424cd scripts/MODELLER/SEA3 && python all_sjkim_final2.py: SEA3 430-536cd scripts/MODELLER/SEA3 && python all_sjkim_final3.py: SEA3 1092-1139cd scripts/MODELLER/SEA4 && python all_sjkim_final1.py: SEA4 45-426cd scripts/MODELLER/SEA4 && python all_sjkim_final2.py: SEA4 659-835cd scripts/MODELLER/SEA4 && python all_sjkim_final3.py: SEA4 942-1032
To produce a single model with 1:3 stoichiometry and rotational
symmetry, as in the publication, use the run_qsub.sh script:
mkdir modeling
cd modeling
../run_qsub.sh 50000 20000 3 True
This script in turn runs scripts/sj_SEA_multi_layers.py, which if desired
can be run with different options to explore other representations and sampling
options.
For the publication, this script was run 885 times to generate the final
ensemble, which was then clustered to produce the 340 models in the output
directory.
Author(s): Seung Joong Kim, Riccardo Pellarin, and Peter Cimermancic
Date: October 6th, 2014
License: LGPL. This library is free software; you can redistribute it and/or modify it under the terms of the GNU Lesser General Public License as published by the Free Software Foundation; either version 2 of the License, or (at your option) any later version.
Testable: Yes.
Parallelizeable: No
Publications:
- Romain Algret, Javier Fernandez-Martinez, Yi Shi, Seung Joong Kim, Riccardo Pellarin, Peter Cimermancic, Emilie Cochet, Andrej Sali, Brian T. Chait, Michael P. Rout, and Svetlana Dokudovskaya, Molecular architecture and function of the SEA complex, a modulator of the TORC1 pathway, Molecular & Cellular Proteomics, 2014, mcp.M114.039388.



 Download files
Download files Verified to work with the
Verified to work with the  To install the software needed to reproduce this system with the
To install the software needed to reproduce this system with the
 To set up the environment on the UCSF Wynton cluster to run
this system, run:
To set up the environment on the UCSF Wynton cluster to run
this system, run: